
Takara Bio USA, Inc.
2560 Orchard Parkway, San Jose, CA 95131, USA
U.S. Technical Support: technical_support@takarabio.com
United States/Canada
800.662.2566
Asia Pacific
+1.650.919.7300
Europe
+33.(0)1.3904.6880
Japan
+81.(0)77.565.6999
Page 1 of 41
Takara Bio USA
Matchmaker® Gold
Yeast Two-Hybrid
System User Manual
Cat. Nos. 630466, 630489, 630498, 630499
(042424)

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 2 of 41
Table of Contents
I. Introduction & Protocol Overview.................................................................................................................................. 4
II. List of Components ......................................................................................................................................................... 7
III. Additional Materials Required .................................................................................................................................... 8
IV. General Considerations Regarding Yeast Two-Hybrid Libraries ............................................................................... 9
V. List of Abbreviations .................................................................................................................................................... 10
VI. Control Experiments ................................................................................................................................................. 11
A. General Considerations ............................................................................................................................................. 11
B. Protocol: Control Mating Protocol ............................................................................................................................ 12
VII. Cloning and Testing Bait for Autoactivation and Toxicity ....................................................................................... 14
A. Generate a Bait Clone ............................................................................................................................................... 14
B. Detecting Bait Expression ......................................................................................................................................... 15
C. Protocol: Testing Your Bait for Autoactivation ........................................................................................................ 15
D. Protocol: Testing Your Bait for Toxicity .................................................................................................................. 16
VIII. Two-Hybrid Library Screening Using Yeast Mating ................................................................................................ 17
IX. Analysis of Results.................................................................................................................................................... 21
X. Confirmation of Positive Interactions & Rescue of the Prey Plasmid .......................................................................... 22
A. Protocol: Yeast Colony PCR Analysis to Eliminate Duplicate Clones .................................................................... 23
B. Protocol: Rescue and Isolation of Library Plasmid Responsible for Activation of Reporters .................................. 23
C. Protocol: Distinguishing Genuine Positive from False Positive Interactions ........................................................... 24
D. Sequence Analysis of a Genuine Positive ................................................................................................................. 26
E. Biochemical Methods to Confirm Positive Interactions ........................................................................................... 27
F. Downstream Analysis ............................................................................................................................................... 27
XI. Troubleshooting Guide ............................................................................................................................................. 28
XII. References ................................................................................................................................................................. 30
Appendix A: Mate & Plate Library Construction ................................................................................................................. 32
Appendix B: Library Titering ............................................................................................................................................... 35
Appendix D: Yeast Growth Media & Supplements .............................................................................................................. 37

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 3 of 41
Table of Figures
Figure 1. The two-hybrid principle. ........................................................................................................................................ 4
Figure 2. Reporter gene constructs in Matchmaker yeast strains. ........................................................................................... 5
Figure 3. Two-hybrid screening using Mate & Plate Libraries............................................................................................... 6
Figure 4. Simple, one-step PCR cloning with In-Fusion HD Cloning Plus Kits. ................................................................. 14
Figure 5. An example of a typical yeast zygote. ................................................................................................................... 18
Figure 6. High stringency screening of potential interactors. ............................................................................................... 20
Figure 7. Strategies for analyzing and verifying putative positive interactions. ................................................................... 22
Figure 8. Illustration of the activation of reporter gene expression in genuine and false positives. ..................................... 24
Figure 9. Using cotransformation on selective media to verify protein interactions ............................................................ 25
Figure 10. Map of pGBKT7-53 DNA-BD control plasmid. ................................................................................................. 36
Figure 11. Map of pGADT7-T AD control plasmid. ............................................................................................................ 36
Table of Tables
Table I. Mating the Pretransformed Control Strains ............................................................................................................. 11
Table II. Yeast Host Strain Genotypes .................................................................................................................................. 32
Table III. Phenotype Testing on Various SD Media ............................................................................................................. 32
Table IV. Components of Yeast Media Set 2 & Yeast Media Set 2 Plus ............................................................................. 37
Table V. Individual Yeast Media Pouches for Matchmaker Gold Protocols ........................................................................ 38
Table VI. Additional Media & Media Supplements Required for a Two-Hybrid Screen .................................................... 38
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 4 of 41
I. Introduction & Protocol Overview
Principle of the Two-Hybrid Assay
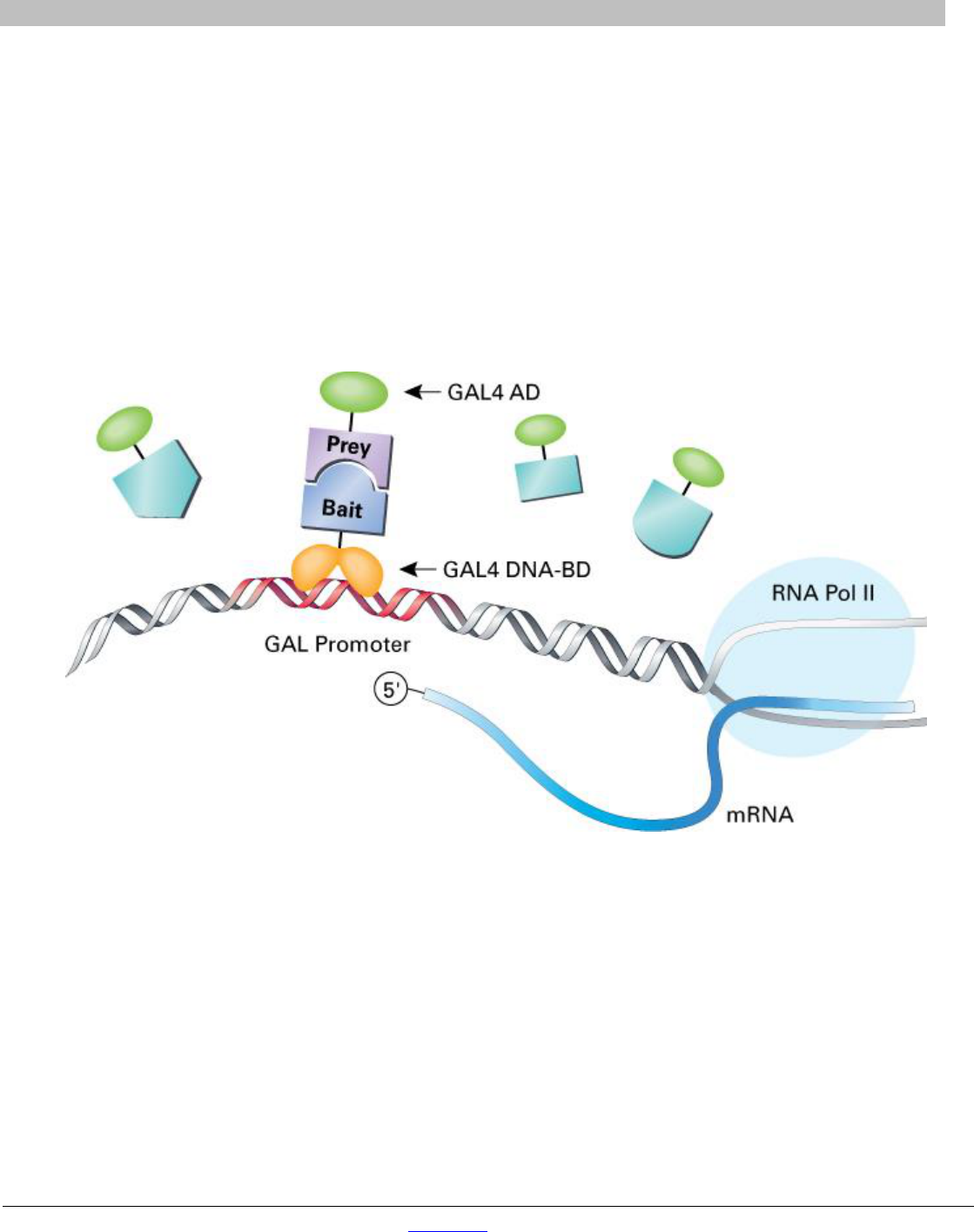
In a Matchmaker GAL4-based two-hybrid assay, a bait protein is expressed as a fusion to the Gal4 DNA-binding
domain (DNA-BD), while libraries of prey proteins are expressed as fusions to the Gal4 activation domain (AD;
Fields & Song, 1989; Chien et al. 1991). In the Matchmaker Gold Yeast Two-Hybrid System (Cat. No.
630489), when bait and library (prey) fusion proteins interact, the DNA-BD and AD are brought into proximity to
activate transcription of four independent reporter genes (AUR1-C, ADE2, HIS3, and MEL1) (Figure 1).
This technology can be used to:
• identify novel protein interactions
• confirm putative interactions
• define interacting domains
Figure 1. The two-hybrid principle. Two proteins are expressed separately, one (a bait protein) fused to the Gal4 DNA-
binding domain (BD) and the other (a prey protein) fused to the Gal4 transcriptional activation domain (AD). In yeast strain
Y2HGold, activation of the reporters (AUR1-C, ADE2, HIS3, and MEL1) only occurs in a cell that contains proteins which interact and
bind to the Gal4-responsive promoter.
The Bait
To make your GAL4 DNA-BD/bait construct, we recommend using pGBKT7, which is available separately (Cat.
No. 630443) or as a component of our Matchmaker Gold Two-Hybrid System (Cat. No. 630489). To investigate
ternary protein complexes, we suggest you use pBridge (Cat. No. 630404), a three-hybrid vector that contains two
MCS regions so that you can express a Gal4 DNA-BD fusion and a second protein of interest that may act as a
"bridge" between bait and prey.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 5 of 41
Four Reporter Genes to Detect Protein Interactions
There are four integrated reporter genes under the control of three distinct Gal4-responsive promoters (Figure 2)
in Takara Bio’s Y2HGold Yeast Strain, which are used to detect two-hybrid interactions.
AUR1-C. A dominant mutant version of the AUR1 gene that encodes the enzyme inositol phosphoryl ceramide
synthase. AUR1-C is expressed in Y2HGold Yeast Strain in response to protein-protein interactions that bring the
GAL4 transcriptional activation and DNA binding domains into close proximity. In Saccharomyces cerevisiae, its
expression confers strong resistance (AbA
r
) to the otherwise highly toxic drug Aureobasidin A (Cat Nos. 630466
& 630499). This drug reporter is preferable to nutritional reporters alone, due to lower background
activity. For example, the use of this reporter alone results in considerably less background than a histidine
reporter alone.
HIS3. Y2HGold is unable to synthesize histidine and is therefore unable to grow on media that lack this essential
amino acid. When bait and prey proteins interact, Gal4-responsive His3 expression permits the cell to
biosynthesize histidine and grow on –His minimal medium.
ADE2. Y2HGold is also unable to grow on minimal media that does not contain adenine. However, when two
proteins interact, Ade2 expression is activated, allowing these cells to grow on –Ade minimal medium.
MEL1. MEL-1 encodes a-galactosidase, an enzyme occurring naturally in many yeast strains. As a result of two-
hybrid interactions, a-galactosidase (MEL1) is expressed and secreted by the yeast cells. Yeast colonies that
express Mel1 turn blue in the presence of the chromogenic substrate X-alpha-Gal (Cat. Nos. 630462 & 630462).
NOTE: X-alpha-Gal is not X-Gal and is not a substrate for β-galactosidase.
Three Different Binding Sites
Three promoters controlling the four reporter genes AUR1-C, HIS3, ADE2, and MEL1 in Y2HGold are unrelated
except for the short protein binding sites in the UAS region that are specifically bound by the Gal4 DNA-BD.
Thus, library proteins that interact with unrelated sequences flanking or within the UAS (i.e., false positives) are
automatically screened out.
Figure 2. Reporter gene constructs in Matchmaker yeast strains. In Y2HGold, the HIS3, ADE2, and MEL1/AUR1-C reporter genes are under the
control of three completely heterologous Gal4-responsive promoter elements—G1, G2, and M1, respectively. The protein-binding sites within the
promoters are different, although each is related to the 17-mer consensus sequence recognized by Gal4 (Giniger et al. 1985; Giniger & Ptashne,
1988).
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 6 of 41
Matchmaker Screening Protocol Overview
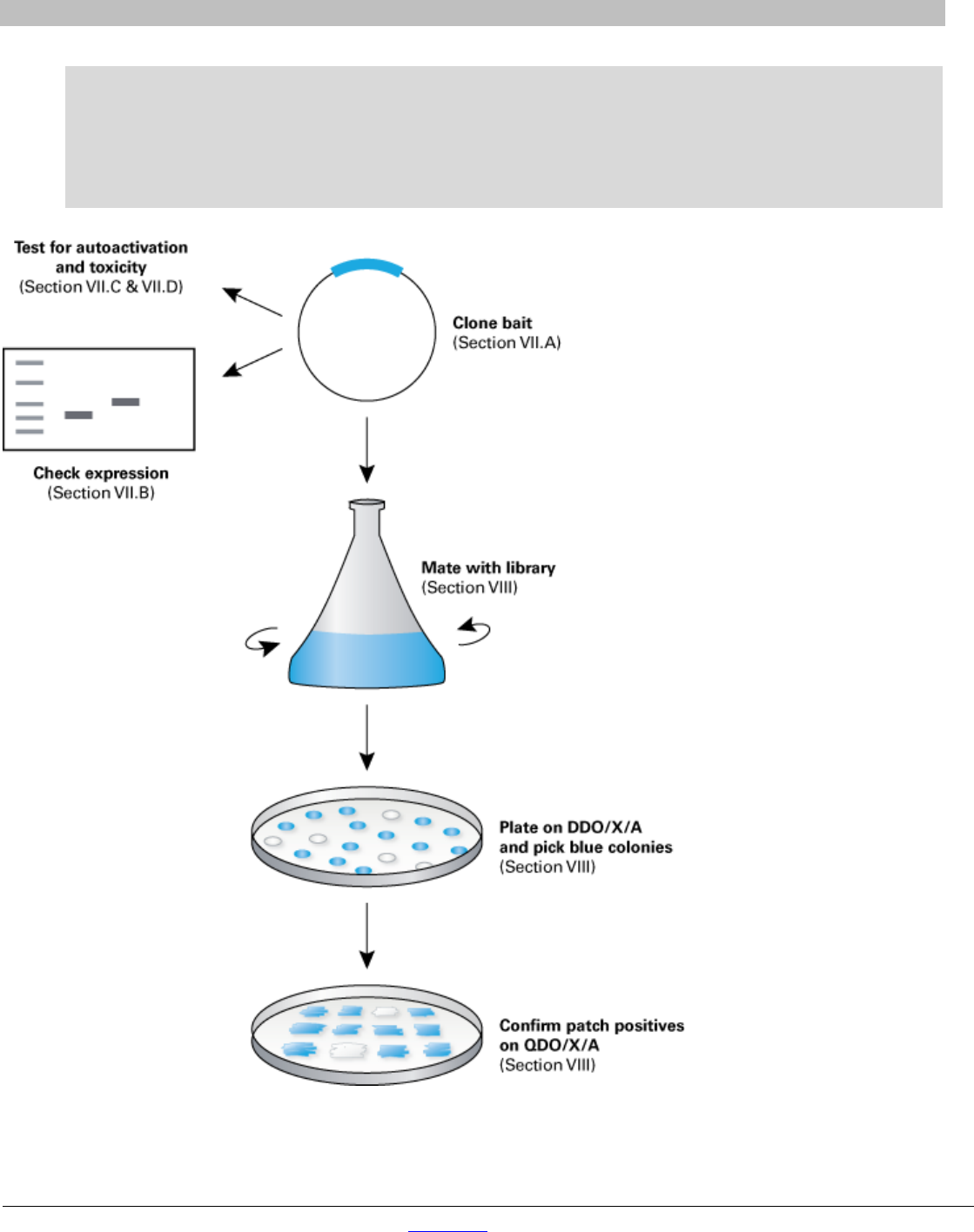
The entire Matchmaker screening process (Figure 3) consists of the following steps:
• Step 1. Perform control experiments
• Step 2. Clone and test bait for autoactivation and toxicity
• Step 3. Screen Mate & Plate library
• Step 4. Confirm and interpret results
Figure 3. Two-hybrid screening using Mate & Plate Libraries. Your bait protein is expressed as a fusion with the Gal4 DNA-BD in yeast strain
Y2HGold. The high-complexity library, which expresses fusions with the Gal4 AD, is provided in yeast strain Y187. When cultures of the two
transformed strains are mixed together overnight, they mate to create diploids. Diploid cells contain four reporter genes: HIS3, ADE2, MEL1, and
AUR1-C, that are activated in response to two-hybrid interactions.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 7 of 41
II. List of Components
Matchmaker Gold Yeast Two-Hybrid System (Cat. No. 630489)
Store all Matchmaker vectors at –20°C, all yeast (S. cerevisiae) strains at –70°C, and all Yeast Media Pouches at
room temperature.
For the Yeastmaker™ Yeast Transformation System 2, store carrier DNA and control plasmid at –20°C, and all
other components at room temperature.
Matchmaker Vectors
•
50 µl
pGBKT7 DNA-BD Cloning Vector (0.1 µg/µl)
•
50 µl
pGADT7 AD Cloning Vector (0.1 µg/µl)
•
50 µl
pGBKT7-53 Control Vector (0.1 µg/µl)
•
50 µl
pGADT7-T Control Vector (0.1 µg/µl)
•
50 µl
pGBKT7-Lam Control Vector (0.1 µg/µl)
Matchmaker Yeast Strains
•
0.5 ml
Y2HGold Yeast Strain
•
0.5 ml
Y187 Yeast Strain
Yeastmaker Yeast Transformation System 2 (also available separately as Cat No. 630439)
•
2 x 1 ml
Yeastmaker Carrier DNA, denatured (10 mg/ml)
•
20 µl
pGBT9 (0.1 µg/µl; positive control plasmid)
•
2 x 50 ml
50% PEG
•
50 ml
1 M LiAc (10X)
•
50 ml
10X TE Buffer
•
50 ml
YPD Plus Liquid Medium
Complimentary Yeast Media Pouches
•
1 x 0.5 L
YPDA
•
1 x 0.5 L
YPDA with Agar
•
1 x 0.5 L
SD–Leu with Agar
•
1 x 0.5 L
SD–Trp with Agar

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 8 of 41
III. Additional Materials Required
The following reagents are required but not supplied.
A. Mate & Plate Libraries
(several catalog items; see www.takarabio.com for current list)
Store all Mate and Plate Libraries and Control vials at –70°C.
5 x 1.0 ml Library Aliquots (Universal Human and Mouse are also sold as 2 x 1 ml vials)
1 x 1.0 ml Mate & Plate Control
Alternatively, you can make your own Mate & Plate library using our Make Your Own “Mate & Plate”
Library System (Cat. No. 630490).
B. Accessory Kits
• Yeastmaker Yeast Transformation System 2 (supplied with your system—see Section II; also sold
separately as Cat. No. 630439)
• Easy Yeast Plasmid Isolation Kit (50 preps; Cat. No. 630467)
• Matchmaker Insert Check PCR Mix 2 (Cat. No. 630497; for characterizing the cDNA inserts of
positive clones from your library screening.)
C. Tools for Plating Yeast
Tools for plating yeast include a sterile glass rod—and a bent Pasteur pipette or 5-mm glass beads for
spreading cells on plates. (Use 5–7 beads per 100-mm plate, or 15–20 beads for a 150-cm plate).
D. Yeast Media
Table IV (in Appendix D) lists the components of the Yeast Media Set 2 (Cat. No. 630494) and the Yeast
Media Set 2 Plus (Cat. No. 630495). These media sets contain a complete assortment of mixes for
preparing eight specialized broth and agar media, designed for use with the Matchmaker Gold Yeast Two-
Hybrid System, in convenient, “ready-mixed” foil pouches. The Yeast Media Set 2 Plus also contains the
additional media supplements Aureobasidin A and X--Gal, which are required for the protocols
described in this user manual. Table V (in Appendix D) contains information for purchasing each of the
media mixes separately, in packs of 10 pouches, and Table VI (in Appendix D) contains preparation
instructions for all additional required media supplements and information for purchasing Aureobasidin A
and X--Gal separately.
Additionally, the following should be considered when culturing yeast for a two-hybrid screen:
• See Appendix D for working and stock concentrations of Aureobasidin A and X--Gal.
• SD medium (synthetically defined medium) is minimal media that is routinely used for culturing
S. cerevisiae. SD base supplies everything that a yeast cell needs to survive (including carbon
and nitrogen sources). Essential amino acids, which are added to SD base to create minimal
medium, are already included premixed in Takara Bio’s Yeast Media Pouches. The particular
minimal medium that is chosen will determine which plasmids and/or activated reporters are
selected for.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 9 of 41
• SD/–Leu/–Trp dropout supplement is used to select for the bait and prey plasmids. SD/–Leu/–
Trp dropout is so called because the medium includes every essential amino acid except for
leucine and tryptophan, which are omitted from the formulation (or “Dropped Out”). Cells
harboring Matchmaker bait and prey plasmids are able to grow because the plasmids encode
tryptophan and leucine biosynthesis genes, respectively, that are otherwise absent from the cell.
We often refer to SD/–Leu/–Trp as Double Dropout (DDO) in this user manual.
• SD/–Ade/–His/–Leu/–Trp dropout supplement is used to select for the bait and prey plasmids,
and in addition, for the activation of the Gal-responsive HIS3 and ADE2 genes as part of the
confirmation step of the two-hybrid assay. Colonies that grow on this Quadruple Dropout
(QDO) contain both bait and prey plasmids and also express proteins that interact with each other
to activate HIS3 and ADE2. This medium is used at the end of the two-hybrid screen to confirm
interactions.
IV. General Considerations Regarding Yeast Two-Hybrid Libraries
Use of Mate & Plate Libraries
Please note that the protocols described in this manual assume that you are using a Mate & Plate Library. These
libraries utilize the natural ability of haploid yeast strains such as Y187 and Y2HGold to mate with each other to
form a diploid cell, providing a very easy way to introduce an entire library (prey) to your bait.
• Takara Bio strongly recommends using Mate & Plate Libraries with the Matchmaker Gold System.
These libraries provide by far the simplest method for yeast two-hybrid screening because no library-scale
transformations or labor-intensive amplifications are needed. Thus, very little optimization and hands-on
time are required.
• Several Mate & Plate libraries are available for purchase from Takara Bio, supplied as 5 x 1 ml vials (and
also sold as 2 x 1 ml vials for Universal Human and Universal Mouse libraries). Alternatively, you can
easily “Make Your Own Mate & Plate Library” using Cat No. 630490, and store enough 1 ml vials for
more than 100 library screens.
• A single 1-ml Mate & Plate Library aliquot is sufficient for each complete library screening (>1 x 10
6
independent clones).
• See Appendix A for more details on prey vectors used to construct Mate & Plate Libraries. These libraries
are supplied in Saccharomyces cerevisiae strain Y187, in Freezing Medium. Depending on which library
you purchase, your library may be cloned into pGADT7-Rec2, pGADT7-RecAB, or pACT2. (See
Certificate of Analysis for details.). All are compatible with Matchmaker Gold.
• A Mate & Plate Control is supplied with all Mate & Plate libraries. This control is Y187 Yeast Strain
pretransformed with our pGADT7-T positive control plasmid, which expresses the Gal4 AD-SV40 large
T-antigen fusion protein. See Section VI for control experiments.
• Once a library aliquot has been thawed, do not refreeze it. With every freeze/thaw cycle, there is a ~10%
loss in viability, which can affect the quality of the library.
• The recommended Freezing Medium consists of YPDA broth + 25% glycerol (see Appendix D).

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 10 of 41
V. List of Abbreviations
AD fusion library
[or AD library]
A cDNA library (such as a Mate & Plate Library) constructed in an activation domain
(AD) vector such that the proteins encoded by the inserts are fused to the 3’ end of the
Gal4 AD
AD/library plasmid
Plasmid encoding a fusion of the Gal4 activation domain and a library cDNA
AD/library protein
A protein fusion comprised of the Gal4 activation domain and a polypeptide encoded by a
library cDNA
AD vector
Plasmid encoding the yeast Gal4 activation domain
DNA-BD vector
Plasmid encoding the Gal4 DNA-binding domain
DNA-BD/bait
plasmid
Plasmid encoding a fusion of the Gal4 DNA binding domain and a bait cDNA
DNA-BD/bait
protein
[or “bait”]
A protein fusion comprised of the Gal4 DNA binding domain and a polypeptide encoded
by a bait cDNA
Yeast Phenotypes
AbA
The antibiotic Aureobasidin A, which is toxic to yeast at low concentrations (0.1–0.5
µg/ml). It acts by inhibiting a yeast enzyme, inositol phosphoryl ceramide synthase.
AbA
r
Resistance to the antibiotic Aureobasidin A, conveyed by expression of the AUR1-C gene
product
AUR1-C
A dominant mutant version of the AUR1 gene, which encodes the enzyme inositol
phosphoryl ceramide synthase. This gene is expressed in yeast strain Y2HGold, in
response to protein-protein interactions that bring the GAL4 activation and binding
domains in proximity, thus conferring resistance to the antibiotic Aureobasidin A.
Ade–, or His–, or
Leu–, or Trp–
Requires adenine (Ade), or histidine (His) or leucine (Leu), or tryptophan (Trp) in the
medium to grow; i.e., is auxotrophic for one (or more) of these specific nutrients
LacZ
+
Expresses the LacZ reporter gene; i.e., is positive for ß-galactosidase (beta-gal) activity
Mel1
+
Expresses the MEL1 reporter gene; i.e., is positive for α-galactosidase (alpha-gal) activity
Miscellaneous
SD
Minimal, synthetically defined medium for yeast; is comprised of a nitrogen base, a
carbon source (glucose unless stated otherwise), and a DO supplement
DO
Dropout (supplement or solution); a mixture of specific amino acids and nucleosides used
to supplement SD base to make SD medium; DO solutions are missing one or more of the
nutrients required by untransformed yeast to grow on SD medium
DDO
Double dropout medium: SD/–Leu/–Trp
DDO/X/A
Double dropout medium: SD/–Leu/–Trp supplemented with X-alpha-Gal and
Aureobasidin A
TDO
Triple dropout medium: SD/–His/–Leu/–Trp or SD/–Ade/–Leu/–Trp
QDO
Quadruple dropout medium: SD/–Ade/–His/–Leu/–Trp
QDO/X/A
Quadruple dropout medium: SD/–Ade/–His/–Leu/–Trp supplemented with X-alpha-Gal
and Aureobasidin A
YPD
A blend of yeast extract, peptone, and dextrose in optimal proportions for growth of most
strains of S. cerevisiae
YPDA
YPD medium supplemented with adenine hemisulfate (1X concentration = 120 µg/ml)
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 11 of 41
VI. Control Experiments
PLEASE READ THE ENTIRE PROTOCOL BEFORE STARTING. Use this procedure to perform a control
mating before screening a two-hybrid library.
A. General Considerations
To familiarize yourself with the procedures and expected results of a two-hybrid assay, use these
procedures to perform a control mating before you begin screening the library. Select for diploids and for
two-hybrid protein-protein interactors as described below.
• pGBKT7-53 encodes the Gal4 DNA-BD fused with murine p53; pGADT7-T encodes the Gal4
AD fused with SV40 large T-antigen. Since p53 and large T-antigen are known to interact in a
yeast two-hybrid assay (Li & Fields, 1993; Iwabuchi et al. 1993), mating Y2HGold [pGBKT7-
53] with Y187 [pGADT7-T] will result in diploid cells containing both plasmids that can activate
all four reporters (Table I).
• A negative control should also be performed using pGBKT7-Lam (which encodes the Gal4 BD
fused with lamin) and pGADT7-T. Diploid yeast containing pGBKT7-Lam and pGADT7-T will
grow on SD/–Leu, SD/–Trp and SD/–Leu/–Trp (DDO) minimal media, but no colonies should
grow on DDO + AbA.
• Table I indicates the selection media required for strains containing a DNA-BD vector, AD
vector, or both, as well as the selection for diploids expressing interacting proteins.
Table I. Mating the Pretransformed Control Strains
Mating Strain
[plasmid]
Plate on SD Minimal Agar
Medium
Selects for
MEL1
Phenotype
Y2HGold [pGBKT7-
53]
x
Y187[pGADT7-T]
–Leu
pGADT7-T
White
–Trp
pGBKT7-53
White
–Leu/–Trp
1
(DDO)
Diploids containing pGBKT7-53
and pGADT7-T
White
–Leu/–Trp/X-alpha-Gal/AbA
2
(DDO/X/A)
Diploids that have also activated
Aureobasidin A resistance and
a-galactosidase through protein-
protein interactions
Blue
1
Controls for mating efficiency.
2
Selects for diploids expressing interacting proteins.
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 12 of 41
B. Protocol: Control Mating Protocol
1. Materials:
• SD/–Trp with Agar (see Appendix D)
• SD/–Leu with Agar (see Appendix D)
• SD/–Trp/X-alpha-Gal agar plates (see Appendix D)
• SD/–Leu/–Trp agar plates (see Appendix D)
• SD/–Leu/–Trp/X-alpha-Gal /AbA agar plates (see Appendix D)
• 2 x YPDA Broth (see Appendix D)
• YPDA Broth + 25% glycerol (Freezing Medium; see Appendix D)
• Y2HGold Yeast Strain (Bait Strain)
• Y187 Yeast Strain (Prey Strain)
• pGBKT7-53 Positive Control Bait Plasmid
• pGBKT7-Lam Negative Control Bait Plasmid
• pGADT7-T Positive Control Prey Plasmid
NOTES:
• Use the Yeastmaker Yeast Transformation System 2 (supplied with this system) for all
transformations.
• X-alpha-Gal is not the same as X-Gal.
2. Use the Yeastmaker Yeast Transformation System 2 according to the small-scale protocol in the
accompanying user manual to perform the following three transformations.
Strain
Transformation Plasmid
Plating Medium
Y2HGold
pGBKT7-53
SD/–Trp with Agar
Y2HGold
pGBKT7-Lam
SD/–Trp with Agar
Y187
pGADT7-T
SD/–Leu with Agar
3. Grow at 30°C for 3 days.
NOTE: If you wish, you may stop the experiment at this step and resume work later. The plates
can be stored at 4ºC in subdued lighting for up to one month.
4. Pick one 2–3 mm colony of each type for use with the following small-scale mating procedure
(Steps 5–7).
• Positive Control Mating: Y2HGold [pGBKT7-53] and Y187 [pGADT7-T]
• Negative Control Mating: Y2HGold [pGBKT7-Lam] and Y187 [pGADT7-T]
5. Place both colonies in a single 1.5-ml centrifuge tube containing 500 µl of 2X YPDA and vortex
to mix.
6. Incubate with shaking at 200 rpm at 30°C overnight [20–24 hr].
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 13 of 41
7. From the mated culture (0.5 ml), spread 100 µl of 1/10, 1/100, and 1/1,000 dilutions on each of
the following agar plates. Incubate plates (colony side facing downward) at 30°C for 3–5 days.
• SD/–Trp
• SD/–Leu
• SD/–Leu/–Trp (=DDO)
• SD/–Leu/–Trp/X-alpha-Gal/AbA (=DDO/X/A)
8. Expected results after 3–5 days:
Positive control:
• Similar number of colonies on DDO and DDO/X/A agar plates
• Colonies on DDO/X/A are blue
Negative control:
• Colonies on DDO, but no colonies on DDO/X/A agar plates
NOTES:
• For positive interactions, theoretically, the number of colonies should be the same on both
media. DDO selects for the presence of both plasmids (i.e., proper mated diploids) and
DDO/X/A selects for the plasmids as well as for the interactions of the hybrid proteins
encoded by them to activate the AbA
r
and MEL1 reporters. However, a difference
(approximately 10–20% lower on DDO/X/A) is usually observed.
• If you see no colonies on DDO, compare to colony counts on SD/–Trp and SD–/Leu single
dropout media to determine if there was a problem with the bait or the prey cultures,
respectively.
9. Pick healthy 2-mm colonies from DDO plates, restreak onto fresh DDO plates, and incubate at
30°C for 3–4 days.
• Short-term storage (<4 weeks): Seal with Parafilm and store at 4°C.
• Long-term storage: Scoop a large healthy colony and fully resuspend in 500 µl of YPDA
Broth + 25% glycerol (Appendix D). Store at –80°C.
NOTES:
• These diploids are useful as reference strains for checking new batches of growth media, and
for comparisons in future experiments.
• When reviving frozen stocks, remember to restreak onto DDO selective medium.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 14 of 41
VII. Cloning and Testing Bait for Autoactivation and Toxicity
PLEASE READ THE ENTIRE PROTOCOL BEFORE STARTING. Detailed instructions are provided to
test your bait for autoactivation (Section C) and toxicity (Section D).
A. Generate a Bait Clone
Generate a GAL4 DNA-BD fusion by cloning your gene of interest in frame with the GAL4 DNA binding
domain of the bait plasmid pGBKT7 (see Appendix C and www.takarabio.com for map).
For an extremely simple cloning procedure, we recommend using one of Takara Bio’s In-Fusion® HD
Cloning Plus Kits (see www.takarabio.com for details).
Figure 4. Simple, one-step PCR cloning with In-Fusion HD Cloning Plus Kits.
PCR Cloning of Your Bait into pGBKT7
The following method describes a simple and highly efficient method to clone your gene in-frame with
the GAL4 BD in pGBKT7. The magic of In-Fusion HD Cloning Plus (Cat. Nos. 638909, 638910,
638911 & 638920) means that:
• Your bait is automatically cloned in-frame with the bait.
• Virtually every clone contains your insert.
• It does not matter what sites are present on your bait sequence since for In-Fusion Cloning you
do not digest it.
1. Digest pGBKT7 to completion with BamHI and EcoRI, then spin column-purify.
2. Amplify your bait insert by PCR using oligos that contain a 24-bp homology to your bait, and a
15-bp homology to the linear ends of pGBKT7, which are designed as follows:
Forward Primer (111 = first codon of your bait)
5’-C ATG GAG GCC GAATTC 111 222 333 444 555 666 777 888
Reverse Primer (LLL = reverse complement of last codon of your bait)
5’-GC AGGTCGACGGATCC LLL NNN NNN NNN NNN NNN NNN NNN
NOTE: These primers actually contain 16 bp of homology in order to keep the BamHI and EcoRI
sites intact.
3. Mix the bait and linear pGBKT7 together and “fuse”’ using the In-Fusion enzyme.
See the In-Fusion HD Cloning Plus Kit User Manual at www.takarabio.com for additional details
regarding PCR cloning procedures.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 15 of 41
• If you wish to find proteins that interact with a membrane-bound or secreted protein, it may be
necessary to first modify the protein (van Aelst et al. 1993) or to use only selected domains as the
bait (as in Kuo et al. 1997).
• In order to confirm that the fusion construct is in-frame, the fusion junction may be sequenced
using a standard T7 primer.
B. Detecting Bait Expression
If you wish to determine whether or not your bait is expressed well in yeast, both of the following
antibodies will detect bait proteins in yeast containing pGBKT7-based bait plasmids (via Western blot).
In order to make yeast protein extracts (yeast cannot simply be boiled or sonicated to extract protein), we
strongly recommend that you use the supporting protocols provided at www.takarabio.com
• GAL4 DNA-BD Monoclonal Antibody (Cat. No. 630403)
• c-Myc Monoclonal Antibody (Cat. No. 631206)
NOTE: Use Y2HGold [pGBKT7-53] as a positive control that expresses a 57 kD protein.
C. Protocol: Testing Your Bait for Autoactivation
As a first step for any two-hybrid screen, it is imperative to confirm that your bait does not autonomously
activate the reporter genes in Y2HGold, in the absence of a prey protein.
1. Materials:
• pGBKT7 containing your gene of interest cloned in frame with the GAL4 DNA-BD
(pGBKT7).
• Competent Y2HGold cells [see Yeastmaker Yeast Transformation System 2 User Manual
(PT1172-1), supplied with this system]
• SD/–Trp/X-alpha-Gal agar plates (Appendix D)
• SD/–Trp/X-alpha-Gal/AbA agar plates (Appendix D)
NOTE: X-alpha-Gal is required, not X-Gal (Appendix D).
2. Transform 100 ng of your pGBKT7+Bait construct using the Yeastmaker Yeast Transformation
System 2 (supplied with this system).
NOTE: (POSITIVE and NEGATIVE CONTROLS) For comparison, we recommend that you
also plate the diploid controls that you created in Section VI; but plate them on DDO/X/A since
they contain both bait and prey plasmids).
3. Spread 100 µl of a 1/10 dilution and a 1/100 dilution of your transformation mixture onto
separate plates, as follows:
• SD/–Trp plates = SDO
• SD/–Trp/X-alpha-Gal = SDO/X plates
• SD/–Trp/X-alpha-Gal/AbA = SDO/X/A plates

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 16 of 41
4. Expected results after 3–5 days:
Sample
Selective Agar Plate
Distinct 2-mm Colonies
Color
Bait autoactivation test
SDO
Yes
White
Bait autoactivation test
SDO/X
Yes
White or very
pale blue
Bait autoactivation test
SDO/X/A
No
N/A
Positive control
(Section VI)
DDO/X/A
Yes
Blue
NOTE: If your bait autoactivates the AbA
r
reporter (i.e., blue colonies appear on SDO/X/A),
check to see if it also activates the His3/Ade2 reporters by plating on SD–Trp/–His/–Ade. See
Section XI (Troubleshooting) if your bait activates all the reporters.
D. Protocol: Testing Your Bait for Toxicity
You should demonstrate that your bait protein is not toxic when expressed in yeast. If your bait is toxic to
the yeast cells, both solid and liquid cultures will grow more slowly.
If expression of your bait protein does have toxic effects, you may wish to switch to a vector (such as
pGBT9) that has a lower level of expression.
NOTE: pGBT9 is supplied as a transformation control in Takara Bio's Yeastmaker Transformation
System 2 (supplied with this system).
1. Materials:
• Y2HGold competent cells [see Yeastmaker Yeast Transformation System 2 User Manual
(PT1172-1), supplied with this system]
• SD/–Trp agar plates (Appendix D)
• SD/–Trp broth (Appendix D)
2. Transform 100 ng of the following vectors:
• pGBKT7 (empty)
• pGBKT7 + cloned bait gene
3. Spread 100 µl of 1/10 and 1/100 dilutions of your transformation mixtures onto SD/–Trp.
4. Grow at 30°C for 3–5 days:
NOTE: If your bait is toxic, you may notice that colonies containing your bait vector are
significantly smaller than colonies containing the empty pGBKT7 vector.
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 17 of 41
VIII. Two-Hybrid Library Screening Using Yeast Mating
PLEASE READ THE ENTIRE PROTOCOL BEFORE STARTING. Detailed instructions are provided for
performing a yeast two-hybrid library screening.
• Simply mix a concentrated bait culture with 1 ml of your Mate & Plate library and incubate overnight
before plating on DDO/X/A selective media (see Figure 3).
• In addition to plating on DDO/X/A media, it is imperative that you determine the number of clones that
you screened on DDO (following the protocol below ensures this). Screening fewer than 1 million clones
may result in an inability to detect positive interactions.
With a “normalized” Mate & Plate library, up to threefold fewer clones need to be screened, because gene
representation has been equalized, significantly reducing the abundance of housekeeping genes in the library.
Several normalized libraries are available from www.takarabio.com.
1. Materials:
• Mate & Plate Library—make your own, using our Make Your Own “Mate & Plate” Library
System (Cat. No. 630490) or purchase separately.
• Bait construct transformed into Y2HGold on SD/–Trp (Section VII)
• SD/–Trp Broth (Appendix D)
• 2X YPDA Broth (Appendix D)
• 0.5X YPDA broth (Appendix D)
• kanamycin sulfate (50 mg/ml)
• YPDA + 25% glycerol [Freezing Medium] (Appendix D)
• The following selective SD agar plates (also see Appendix D):
Agar Media
Acronym
Number of Plates
SD/–Trp
—
5–10 (100 mm plates)
SD/–Leu
—
5–10 (100 mm plates)
SD/–Leu/–Trp
DDO
5–10 (100 mm plates)
SD/–Leu/–Trp/X-alpha-Gal/AbA
DDO/X/A
50–55 (150 mm plates)
SD/–Ade/–His/–Leu/–Trp/X-alpha-
Gal/AbA
QDO/X/A
5–10 (100 mm plates)
NOTE: Takara Bio’s Mate & Plate libraries are supplied in yeast strain Y187, so your bait must be in
yeast strain Y2HGold.
If you wish to “Make your Own Mate & Plate Library”, use our kit (Cat. No. 630490)
(see www.takarabio.com for details)
2. Construct your bait, test for autoactivation and toxicity (Section VII).
3. Perform the control experiments (Section VI).

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 18 of 41
NOTE: Control experiments are strongly recommended; the control strains will aid interpretation of
results when you screen your library.
4. Prepare a concentrated overnight culture of the bait strain (Y2HGold [pGBKT7+Bait]) as follows:
a. Inoculate one fresh, large (2–3 mm) colony of your bait strain into 50 ml of SD/–Trp liquid
medium.
b. Incubate shaking (250–270 rpm) at 30°C until the OD
600
reaches 0.8 (16–20 hr).
c. Centrifuge to pellet the cells (1,000 g for 5 min), discard the supernatant.
d. Resuspend the pellet to a cell density of >1 x 10
8
cells per ml in SD/–Trp (4–5 ml).
[The cells can be counted using a hemocytometer.]
5. Combine the Library Strain with the Bait Strain as follows:
a. Thaw a 1-ml aliquot of your library strain in a room temperature water bath. Remove 10 µl for
titering on 100-mm SD/–Leu agar plates (see Appendix B, Section B for library titering
instructions).
NOTE: Use a hemocytometer to count the cells. Your 1 ml library aliquot should contain
>2 x 10
7
cells. To check the titer, spread 100 µl of 1/100, 1/1,000, 1/10,000 dilutions on SD/–Leu
agar plates. If your titer is 2 x 10
7
cells/ml, you will obtain 200 colonies on the 1/10,000 dilution
plate.
b. Combine 1 ml of your Mate & Plate Library with 4–5 ml Bait Strain (from Step 4) in a sterile 2-L
flask.
c. Add 45 ml of 2xYPDA liquid medium (with 50 µg/ml kanamycin).
d. Rinse cells from the library vial twice with 1 ml 2xYPDA and add to the 2-L flask.
6. Incubate at 30°C for 20–24 hr, slowly shaking (30–50 rpm).
IMPORTANT: Use the lowest shaking speed possible that prevents the cells from settling at the base of
the flask. Vigorous shaking can reduce the mating efficiency, but shaking too slowly will cause the cells
to sediment, also lowering the mating efficiency.
7. After 20 hr, check a drop of the culture under a
phase contrast microscope (40X). If zygotes are
present, continue to Step 8, if not, allow mating to
continue; incubate for an additional 4 hr.
NOTE: A zygote typically has a 3-lobed structure
(see Figure 5). The lobes represent the two haploid
parental cells and the budding diploid cell. Some
zygotes may resemble a clover leaf, while others
may take on a shape similar to a “Mickey Mouse”
face.
8. Centrifuge to pellet the cells (1,000 g for 10 min).
Figure 5. An example of a typical yeast zygote.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 19 of 41
9. Meanwhile, rinse the 2-L flask twice with 50 ml 0.5X YPDA (with 50 µg/ml kanamycin), combine the
rinses, and use this to resuspend the pelleted cells.
10. Centrifuge to pellet the cells (1,000g for 10 min) and discard the supernatant.
11. Resuspend all pelleted cells in 10 ml of 0.5X YPDA/Kan liquid medium. Measure the total volume of
cells + medium.
NOTE: e.g., 10 ml medium + 1.5 ml cells = 11.5 ml
12. From the mated culture, spread 100 µl of 1/10, 1/100, 1/1,000, and 1/10,000 dilutions on each of the
following 100-mm agar plates and incubate at 30°C for 3–5 days.
• SD/–Trp
• SD/–Leu
• SD/–Leu/–Trp (DDO)
NOTE: This step is essential to calculate the number of clones screened (see Step 14).
13. Plate the remainder of the culture, 200 µl per 150-mm on DDO/X/A (50–55 plates). Incubate at 30°C for
3–5 days.
14. Calculate the number of screened clones (diploids) by counting the colonies from the DDO plates after
3–5 days.
• Number of Screened Clones = cfu/ml of diploids x resuspension volume (ml)
• It is imperative that at least 1 million diploids are screened, since using less than this will result in
less chance of detecting genuine interactions on Aureobasidin A plates (DDO/X/A).
Example Calculation
• Resuspension volume (Step 11) = 11.5 ml
• Plating Volume = 100 µl
• 50 colonies grew on the 1/1,000 dilution on DDO plates.
Therefore, Number of Clones screened = 50 x 11.5 x 10 x 1,000 = 5.75 million
15. Determine the Mating Efficiency
Mating efficiencies of 2–5% are readily achieved using this procedure. If your mating efficiency is less
than 2% and you cannot screen 1 million diploids (Step 14), refer to the Troubleshooting Guide (Section
XI) for tips on improving the mating efficiency, and screen more clones.
a. Measure viabilities
• No. of cfu/ml on SD/–Leu = viability of the Prey Library
• No. of cfu/ml on SD/–Trp = viability of Bait
• No. of cfu/ml on SD/–Leu/–Trp = viability of diploids
NOTE: The strain (bait or prey) with the lower viability is the "limiting partner."
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 20 of 41
b. Calculate Mating Efficiency (percentage of diploids):
No. of cfu/ml of diploids x 100 = % Diploids
No. of cfu/ml of limiting partner
Example Calculation
• Resuspension volume (Step 11) = 11.5 ml
• Plating Volume = 100 µl
• 5,000 colonies grew on the 1/10,000 on SD/–Trp
• 100 colonies grew on the 1/10,000 dilution on SD/–Leu
• 50 colonies grew on the 1/1,000 dilution on DDO plates
Therefore (in cfu/ml),
- Viability of Prey Library = 1 x 10
7
- Viability of Bait = 5 x 10
8
- Viability of Diploid = 5 x 10
5
Since the Prey Library is the limiting partner in this example, mating efficiency is calculated as
follows:
5 x 10
5
x 100 = 5% Mating Efficiency
1 x 10
7
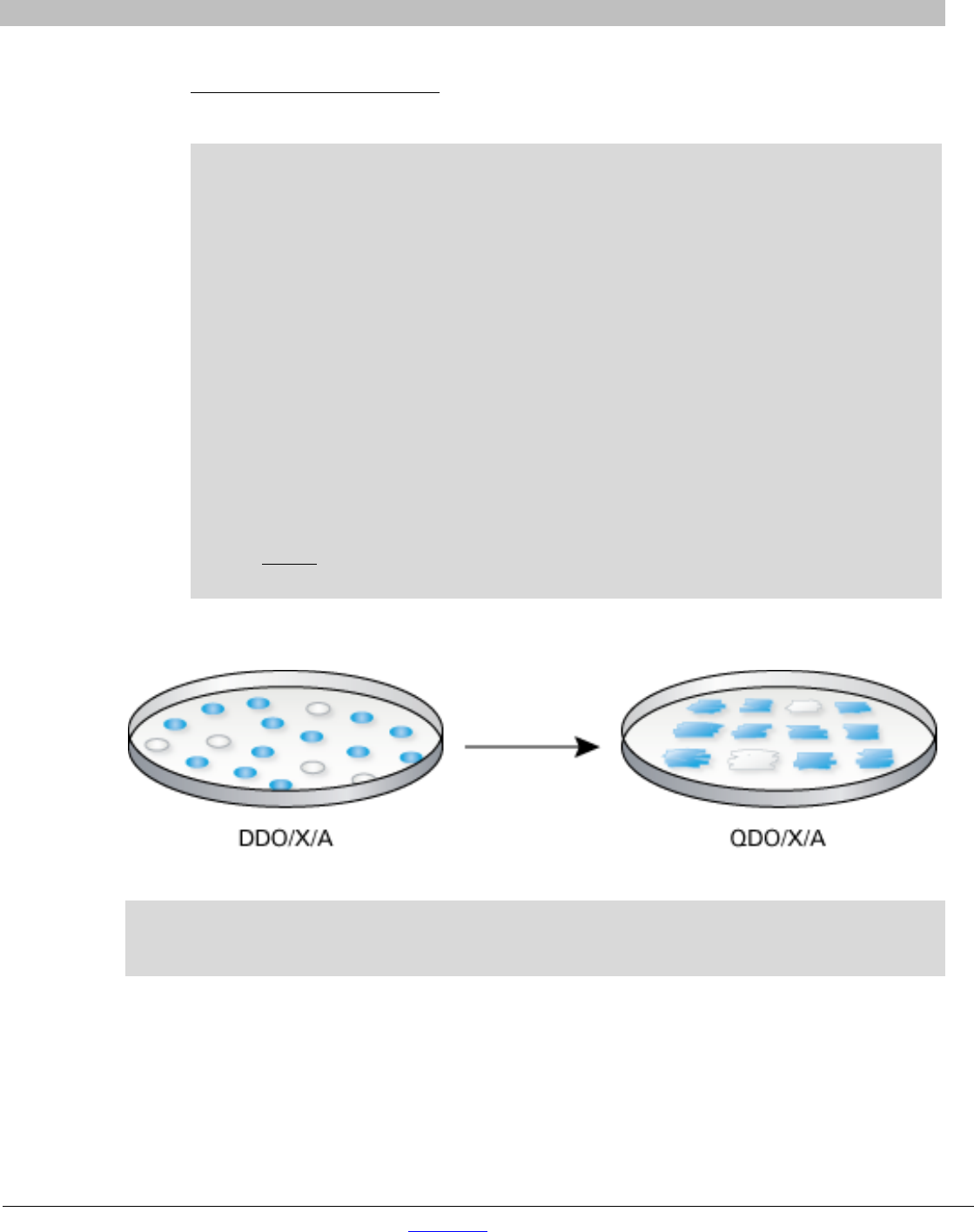
16. Patch out all the blue colonies that grew on DDO/X/A onto higher stringency QDO/X/A agar plates using
a flat sterile toothpick or yellow pipette tip (Figure 6).
Figure 6. High stringency screening of potential interactors.
NOTE: Although it is possible to screen directly on high stringency QDO/X/A, Takara Bio recommends
screening first on lower stringency DDO/X/A to detect as many positives interactors as possible before
confirming those by patching on highest stringency plates.
17. All QDO/X/A positive interactions must be further analyzed (Section X) to identify duplicates and to
verify that the interactions are genuine.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 21 of 41
IX. Analysis of Results
After high-stringency patching to identify potential binding partners for your protein of interest, you may have
very few positives, or too many positives to analyze. In these scenarios, we recommend checking the following:
A. Too Few Positives
Have you screened >1 million independent clones? Refer to Section VIII, Step 14 to determine if you
screened 1 million independent clones? Optimize the mating/transformation procedure (see Section XI.
Troubleshooting Guide) and repeat the screening procedure.
• Check that your DDO/X/A and QDO/X/A growth media performs as expected with the positive
and negative controls. Very little AbA (125 ng/ml) is required, so make certain not to use too
much.
• If you screened >1 million independent clones and detected no positive colonies on DDO/X/A,
repeat the screen with a reduced concentration of Aureobasidin A (150 ng/ml instead of
200 ng/ml).
B. Too Many Positives
Have you determined that your bait does not autoactivate the reporters (Section VII.C)?
• Check that your DDO/X/A and QDO/X/A growth media performs as expected with the positive
and negative controls.
• Your bait may interact with a partner that is abundant in the library. Sort duplicates by Yeast
Colony PCR (Section X.A). After the clones have been sorted into groups, a representative of
each unique type can then be analyzed for false positive interactions (Section X.C).
• Alternatively, you may wish to try a Normalized Mate & Plate Library; see www.takarabio.com
for details.
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 22 of 41
X. Confirmation of Positive Interactions & Rescue of the Prey Plasmid
PLEASE READ THE ENTIRE PROTOCOL BEFORE STARTING. Detailed instructions are provided for
yeast colony PCR to eliminate duplicates (Section A), rescue and isolation of library plasmids responsible for
activation of reporters (Section B), and distinguishing genuine positive from false positive interactions
(Section C).
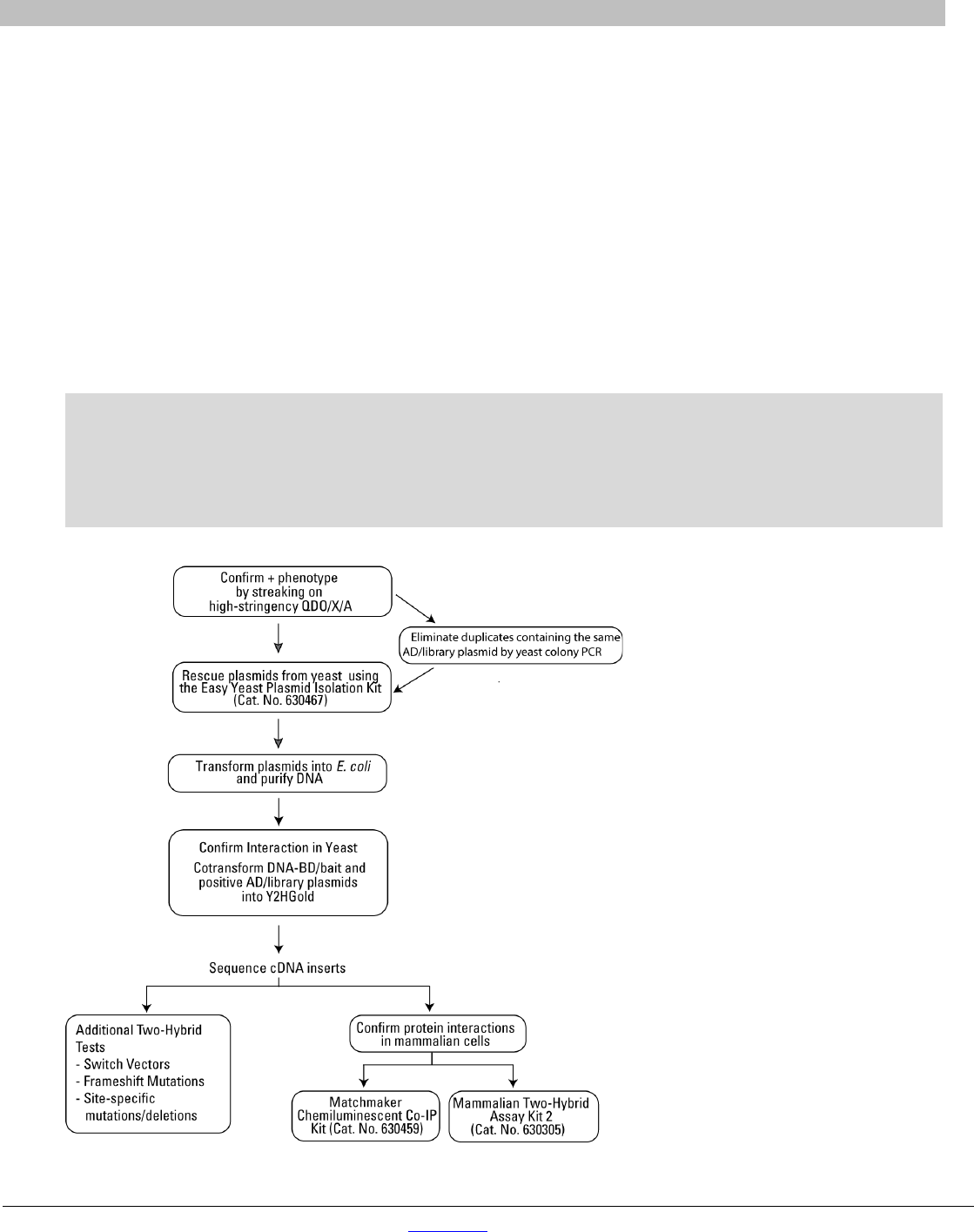
The following represents the recommended order of events to confirm that the positive interactions are genuine.
The strategy is summarized in Figure 7. Note, however, that your preferred order of events may be somewhat
determined by the number of positives obtained from your assay. For instance, if your bait protein interacts with a
protein that is abundant in the library, you may have a large number of potential positives to sort, many of which
may be the same. In this case you may choose to perform colony PCR (Section X.A) to sort the duplicate clones
before segregating and rescuing the plasmid. If you have a low number of positive clones, you may choose to
omit the colony PCR screening step altogether and proceed directly to the Easy Yeast Plasmid Isolation Kit
(Cat. No. 630467).
We recommend performing the following steps prior to sequencing your positive clones:
• Yeast Colony PCR
• Rescue and isolation of the library plasmid responsible for activation of reporters
• Distinguishing genuine positive from false positive interactions
Figure 7. Strategies for analyzing and verifying putative positive interactions.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 23 of 41
A. Protocol: Yeast Colony PCR Analysis to Eliminate Duplicate Clones
1. Use the Matchmaker Insert Check PCR Mix 2 (Cat. No. 630497) to amplify your prey library
inserts. The kit includes a premix of enzyme, reagents, and primers to amplify cDNA inserts from
pGADT7 vectors. You can then characterize the inserts in Steps 2–4 using restriction enzyme analysis
to identify potential duplicates clones. We strongly recommend this complete premix because we find
that it performs very well in yeast colony samples.
2. Analyze PCR products by electrophoresis on a 0.8% TAE Agarose/EtBr gel. The presence of more
than a single band is common, indicating the presence of more than one prey plasmid in a cell.
NOTE: To confirm that similar sized bands contain the same insert, digest the PCR product with
AluI or HaeIII or another frequently cutting enzyme, and analyze the products on a 2% agarose/EtBr
gel.
3. If a high percentage of the colonies appear to contain the same AD/library insert, expand your PCR
analysis to another batch of 50 colonies.
4. At this stage, to quickly identify the clones, the PCR products (observed as a single band on gel) can
be spin column-purified and sequenced using T7 primer.
B. Protocol: Rescue and Isolation of Library Plasmid Responsible for
Activation of Reporters
1. Segregation of Library Plasmid in Yeast
Transformed yeast cells (unlike transformed E. coli cells) can harbor more than one version of a
related plasmid. This means that in addition to containing a prey vector that expresses a protein
responsible for activating the reporters, a yeast cell may also contain one or more prey plasmids that
do not express an interacting protein.
• If you rescue the plasmid via E. coli transformation without first segregating the non-
interacting prey, there is a chance that you will rescue a non-interacting prey plasmid.
• To increase the chance of rescuing the positive prey plasmid, we recommend that you streak
2–3 times on DDO/X (no Aureobasidin A), each time picking a single blue colony for
restreaking. After the first streaking, you may see a mixture of blue and white colonies,
indicating segregation of positive interactors (blue) from non-interactors (white). After
streaking one or two more times, you should only see blue colonies. The plasmid should be
rescued from one of these clones (see Step 2).
2. Rescuing the Library Plasmid from Yeast
The following methods are recommended for rescuing your plasmid from yeast:
• To identify the gene responsible for the positive interaction, rescue the plasmid from yeast
cells grown on QDO/X using the Easy Yeast Plasmid Isolation Kit (Cat. No. 630467)
• If your bait is cloned in pGBKT7 (which contains a kanamycin resistance gene), you can
select for the prey plasmid simply by selection on LB plus 100 µg/ml ampicillin using any
commonly used cloning strain of E. coli, e.g., DH5α, or Stellar™ Competent Cells (Cat. No.
636763).
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 24 of 41
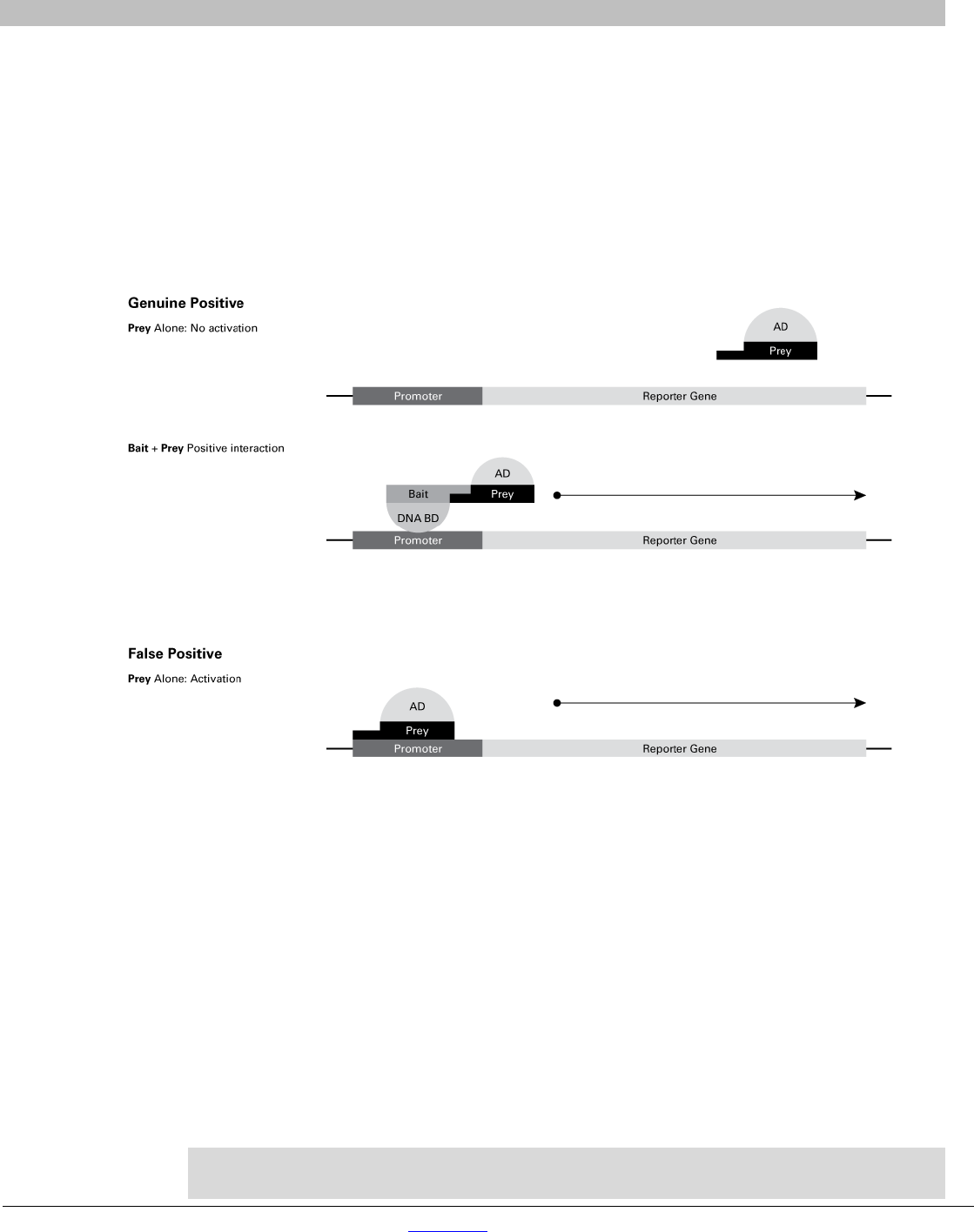
C. Protocol: Distinguishing Genuine Positive from False Positive Interactions
Y2HGold Yeast Strain contains four reporters under the control of three distinct GAL4 UAS sequences.
As a result, of following the high-stringency screening protocols described in this user manual, the
incidence of false positives is reduced to a minimum compared to other systems. The incidence of false
positives is further reduced with Normalized Mate & Plate Libraries due to more equal representation of
each transcript. However, with every two-hybrid screen there is a chance of detecting false positives and
it is important to confirm that your interactions are genuine using the following criteria (see Figure 8):
• Genuine Positive: Both Bait and Prey are required to activate the Gal4-responsive reporters
• False Positive: Prey can activate the Gal4-responsive reporters in the absence of your bait.
Figure 8. Illustration of the activation of reporter gene expression in genuine and false positives.
You can confirm protein interactions in yeast on selective media (see Appendix D for recipes) using the
following cotransformation procedure (Figure 9). This can also be done by yeast mating (see Section
VIII).
1. Materials:
• Competent Y2HGold cells [see Yeastmaker Yeast Transformation System 2 User Manual
(PT1172-1), supplied with this system]
• SD/-Leu/-Trp/X-alpha-Gal Agar (Appendix D) = DDO/X
• SD/-Ade/-His/-Leu/-Trp/X-alpha-Gal/AbA (Appendix D) = QDO/X/A
2. Using the small-scale transformation procedure, cotransform100 ng of each of the following pairs
of vectors into Y2HGold Competent Cells:
• pGBKT7/Bait + Prey (in pGADT7, pGADT7-Rec, or pGADT7-RecAB)
• Empty pGBKT7 + Prey (in pGADT7, pGADT7-Rec, or pGADT7-RecAB)
NOTE: We recommend that you perform the experiment side by side with the positive and
negative controls (Section VI).
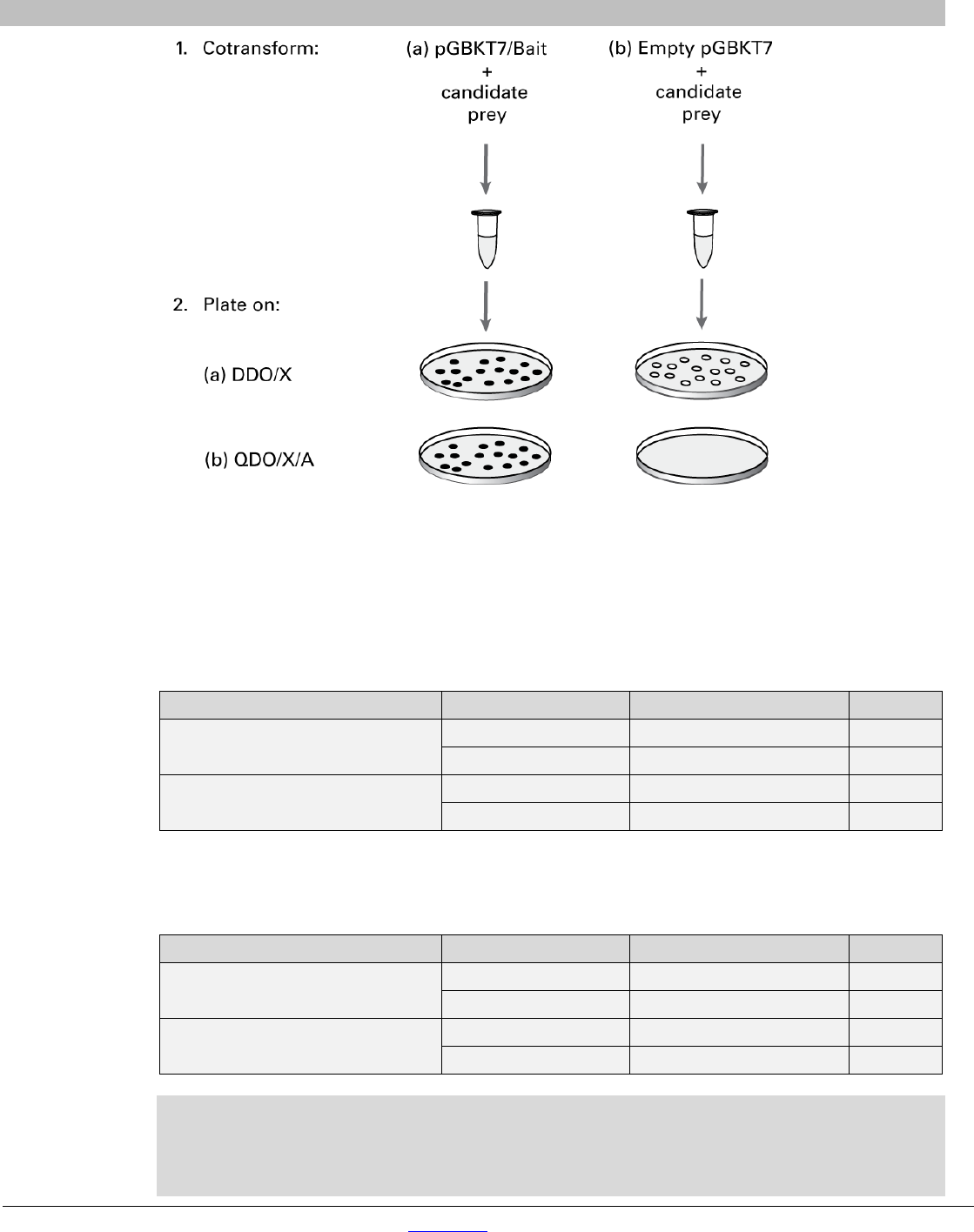
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 25 of 41
Figure 9. Using cotransformation on selective media to verify protein interactions. Expected results from genuine interactions.
3. Spread 100 µl of 1/10 and 1/100 dilutions of the transformation mix on the following plates:
• DDO/X
• QDO/X/A
4. Expected results after 3–5 days at 30°C (Figure 9):
a. Genuine Positive:
Sample
Selective Agar Plate
Distinct 2-mm Colonies
Color
Bait + candidate prey
DDO/X
Yes
Blue
QDO/X/A
Yes
Blue
Empty pGBKT7 + candidate prey
DDO/X
Yes
White
QDO/X/A
No
N/A
b. False Positive:
For false positive interactions, similar numbers of blue colonies are observed on all plates
(indicating that the prey does not require your bait to activate the reporters).
Sample
Selective Agar Plate
Distinct 2-mm Colonies
Color
Bait + candidate prey
DDO/X
Yes
Blue
QDO/X/A
Yes
Blue
Empty pGBKT7 + candidate prey
DDO/X
Yes
Blue
QDO/X/A
Yes
Blue
NOTE: Theoretically, for positive interactions, the number of colonies should be the same on both
media: DDO selects for both plasmids, and QDO/X/A selects for the plasmids as well as for the
interaction of the hybrid proteins encoded by them. However, a difference is usually observed
(10–60% lower on QDO/X/A, depending on the strength of the interaction).

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 26 of 41
D. Sequence Analysis of a Genuine Positive
Once an interaction has been verified as being genuine, the prey insert can be identified by sequencing.
Use only DNA isolated from E. coli for this procedure. AD/library cDNA inserts can be sequenced using
the following:
• Matchmaker AD LD-Insert Screening Amplimer Set (Cat. No. 630433),
• T7 Sequencing Primer
Always check the vector sequence against the primer you wish to use. Be aware that some Matchmaker
AD plasmids (e.g., pACT2) do not contain a T7 Promoter.
Verify the presence of an open reading frame (ORF) fused in frame to the GAL4 AD sequence, and
compare the sequence to those in GenBank, EMBL, or other databases.
NOTES:
Before considering any of the following possibilities, we recommend that you verify that your clone is not
a false positive (Section X.C).
• Most genuine positive clones will activate all reporters, however it is possible that some library
clones only activate a selection of the reporters, for example the colony grows on QDO but does not
turn blue in the presence of X-alpha-Gal. This may be due to inaccessibility of a particular prey
fusion protein to a specific UAS. Confirm the interaction by additional means such as
coimmunoprecipitation.
• Most library clones will contain some 3’ untranslated region, be sure to scan the entire sequence to
find any portion of coding region fused in-frame to the GAL4 AD.
• Yeast tolerate translational frameshifts. A large ORF in the wrong reading frame may correspond to
the protein responsible for the interaction. To verify this, re-clone the insert in-frame (this can be
easily done using Takara Bio’s In-Fusion PCR Cloning Systems (see www.takarabio.com) and
determine if the AbA
r
, ADE2, HIS3, and MEL1 reporters are still active if your bait is also present.
• If your sequencing results reveal a very short peptide (<10 amino acids) fused to the AD—or no
fusion peptide at all—keep sequencing beyond the stop codon. You may find another (larger) open
reading frame (ORF). Such gaps can occur when a portion of the 5' untranslated region of an mRNA
is cloned along with the coding region. A Western blot using Gal4 AD Antibody (Cat. No. 630402)
will reveal the presence and size of an AD fusion protein.
• In some cases, two different ORFs may be expressed as a fusion with the AD even though a non-
translated gap comes between them. This is due to occasional translational read-through.
• If your sequencing results fail to reveal any ORF in frame with the AD coding region, it could be that
the positive library clone is transcribed in the reverse orientation from a cryptic promoter within the
ADH1 terminator on the bait plasmid (Chien et al. 1991).

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 27 of 41
E. Biochemical Methods to Confirm Positive Interactions
We recommend confirming positive interactions using the following methods:
• After sequencing the positive clones, most researchers choose to confirm each protein-protein
interaction using independent, biochemical methods, such as affinity chromatography and/or
immunoprecipitation (Fields & Sternglanz, 1994).
• You can also test protein-protein interactions in mammalian cells using either the Matchmaker
Mammalian Two-Hybrid Assay Kit 2 (Cat. No. 630305) or the pCMV-Myc & pCMV-HA
Vector Set (Cat. No. 631604) The Vector Set includes c-Myc and HA-Tag antibodies for the
isolation and identification of protein-protein complexes.
F. Downstream Analysis
You may wish to compare the strengths of two different interactions—for example, between a bait and
two different prey proteins; or analyze the effects of point mutations on the strength of interaction, using
the following methods:
Quantitative test for interactions: The Gal-responsive LacZ gene (beta-galactosidase) integrated in
Y187 is not secreted (in contrast to alpha-galactosidase encoded by MEL1) and it cannot be used for
blue/white screening on agar plates. However, LacZ is an ideal reporter for quantitative studies of protein-
protein interactions. We recommend the use of yeast strain Y187 for such quantitative studies because the
LacZ promoter in this strain expresses strongly (Y2HGold/Y187 diploids can also be used). Quantitative
LacZ assays are described in supporting Matchmaker protocols at www.takarabio.com.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 28 of 41
XI. Troubleshooting Guide
Problem
Possible Explanation
Solution
DNA-BD/bait
activates reporter
genes
The bait protein has a transcriptional
activation domain. This is especially likely
if the bait protein is a transcription factor
(Ma & Ptashne, 1987; Ruden et al. 1991;
Ruden, 1992). Acidic amphipathic
domains are often responsible for
unwanted transcriptional activation
(Ruden et al. 1991; Ruden, 1992).
Remove the activating domain by creating specific
deletions within the gene. Retest the deletion
constructs for activation. At the amino acid level, a
net negative charge per 10 amino acids is a
minimal AD. Note that such deletions may also
eliminate a potentially interacting domain.
Excessive
background
• Improper media preparation
Remake SD/–Leu/–Trp/X-alpha-Gal/AbA medium.
• 0.5X YPDA medium is too rich for the
resuspension of transformed cells
Use water, TE, or 0.9% NaCl.
Low mating
efficiency
Insufficient number of pretransformed bait
cells in the mating
When you prepare the overnight liquid culture of
the bait strain, be sure to use a large, fresh colony
for the inoculum. After centrifuging and
resuspending the culture, count the cells using a
hemocytometer. The concentration should be
≥1 x 10
8
cells/ml, an ~100-fold excess over the
pretransformed library cells.
One or both of the hybrid proteins is toxic
to yeast
You may be able to genetically engineer the
hybrid protein in a way that will alleviate its toxicity
but still allow the interaction to occur; or use a
DNA-BD or AD vector that expresses lower levels
of the fusion protein (e.g., pBridge or pGBT9).
Bait protein is toxic to the yeast cells
•
In some cases, strains that do not grow well in
liquid culture will grow reasonably well on
agar plates. Resuspend the colony in 1 ml of
SD/–Trp, then spread the cell suspension on
five 100-mm SD/–Trp plates. Incubate the
plates at 30°C until the colonies are confluent.
Scrape the colonies from each plate, pool
them in one tube, and resuspend in a total of
5 ml of 0.5X YPDA. Use the cell suspension
in the normal mating procedure.
•
It may be necessary to perform the mating on
agar plates (Bendixen et al. 1994) or on filters
(Fromont-Racine et al. 1997). Be sure to set
up controls that will allow you to compare the
library mating efficiency with that of your bait
strain mated to Y187[pGADT7-T] and with
that of Y187[pGADT7-T] mated to
Y2HGold[pGBKT7-53].
• Bait proteins may interfere with mating if they
are highly homologous to proteins involved in
yeast mating (e.g., pheromone receptors). If
sequence information on your bait protein is
available, check it for homology to proteins
known to be involved in yeast mating (Schultz
et al. 1995; Pringle et al. 1997). In the rare
case of homology to a pheromone receptor, it
may be necessary to screen the library using
a conventional library-scale yeast
transformation.
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 29 of 41
Problem
Possible Explanation
Solution
Failure to detect
known protein
interactions
•
If one of the following situations is
occurring, it may interfere with the
ability of the two-hybrid proteins to
interact: (1) the hybrid proteins are not
stably expressed in the host cell; (2)
the fused GAL4 domains occlude the
site of interaction; (3) the hybrid protein
folds improperly; or (4) the hybrid
protein cannot be localized to the yeast
nucleus. (See van Aelst et al. [1993] for
one example.)
Construct hybrids containing different domains of
the bait protein. For example, to study proteins
that normally do not localize to the nucleus, it may
be necessary to generate mutant forms of the
protein that can be transported across the nuclear
membrane.
•
Some types of protein interactions may
not be detectable in a GAL4-based
system.
Try using a LexA-based two-hybrid system.
•
Some protein interactions are not
detectable using any type of two-
hybrid assay.
AD/library plasmid
activates all four
reporters
independent of the
DNA-BD/bait
False positives. The AD/library hybrid
activates transcription and binds because
it recognizes the promoter regions of the
reporters.
Refer to Section X for methods to verify protein
interactions; see Serebriiskii et al. (2000) and
Bartel et al. (1993a) for further discussion of false
positives.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 30 of 41
XII. References
• For general reviews on yeast two-hybrid systems, see Allen et al. 1995; Bartel et al. 1993a, 1993b; Bartel & Fields,
(1997); Fields, 1993; Fields & Sternglanz, 1994; Fritz & Green, 1992; Guarente, 1993; Hopkin, 1996; Luban & Goff,
1995; McNabb & Guarente, 1996; Mendelsohn & Brent, 1994.
• An extensive list of Matchmaker System citations can be obtained from our website (
www.takarabio.com
).
• For additional two-hybrid references, see the Golemis lab Web Site (http://www.fccc.edu:80/research/labs/golemis) or
use MedLine (http://www.ncbi.nlm.nih.gov/PubMed/medline.html) and search under key words "two-hybrid.”
Allen, J. B., Wallberg, M. W., Edwards, M. C. & Elledge, S. J. Finding prospective partners in the library: the yeast two-
hybrid system and phage display find a match. TIBS
20
:511–516 (1995).
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. Current Protocols in
Molecular Biology (John Wiley & Sons, Inc.) (1994).
Bartel, P. L. & Fields, S. The Yeast Two-Hybrid System (Oxford University Press, Oxford) (1997).
Bartel, P. L., Chien, C.-T., Sternglanz, R. & Fields, S. Using the two-hybrid system to detect protein-protein interactions.
In Cellular Interactions in Development: A Practical Approach., ed. Hartley, D. A. (Oxford University Press, Oxford) pp.
153–179 (1993a).
Bartel, P. L, Chien, C.-T., Sternglanz, R. & Fields, S. Elimination of false positives that arise in using the two-hybrid system.
BioTechniques
14
:920–924 (1993b).
Bendixen, C., Gangloff, S. & Rothstein, R. A yeast mating-selection scheme for detection of protein-protein interactions.
Nucleic Acids Res.
22
:1778–1779 (1994).
Borson, N. D., Sato, W. L. & Drewes, L. R. A lock-docking oligo(dT) primer for 5’ and 3’ RACE PCR. PCR Methods Appl.
2
:144–148 (1992).
Chien, C. T., Bartel, P. L., Sternglanz, R. & Fields, S. The two-hybrid system: A method to identify and clone genes for
proteins that interact with a protein of interest. Proc. Nat. Acad. Sci. USA
88
:9578–9582 (1991).
Fields, S. The two-hybrid system to detect protein-protein interactions. METHODS: A Companion to Meth. Enzymol.
5:
116–124 (1993).
Fields, S. & Song, O. A novel genetic system to detect protein-protein interactions. Nature
340
: 245–247 (1989).
Fields, S. & Sternglanz, R
.
The two-hybrid system: an assay for protein-protein interactions. Trends Genet.
10
: 286–292
(1994).
Fritz, C. C. & Green, M. R. Fishing for partners. Current Biol.
2
:403–405 (1992).
Fromont-Racine, M., Rain, J.-C. & Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-
hybrid screens. Nature Genetics
16
:277–282 (1997).
Giniger, E., Varnum, S. M. & Ptashne, M. Specific DNA binding GAL4, a positive regulatory protein of yeast. Cell
40
:767–
774 (1985).
Giniger, E. & Ptashne, M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc. Natl. Acad. Sci.
USA
85
:382–386 (1988).
Golemis, E. A., Gyuris, J. & Brent, R. Analysis of protein interactions; and Interaction trap/two-hybrid systems to identify
interacting proteins. In Current Protocols in Molecular Biology (John Wiley & Sons, Inc.), Sections 20.0 and 20.1 (1996).
Guarente, L. Strategies for the identification of interacting proteins. Proc. Natl. Acad. Sci. USA
90
:1639–1641 (1993).
Gubler, U. & Hoffman, B. J. A simple and very efficient method for generating cDNA libraries. Gene
25
:263–269 (1983).
Guthrie, C. & Fink, G. R. Guide to yeast genetics and molecular biology. In Methods in Enzymology (Academic Press, San
Diego)
194
:1–932 (1991).

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 31 of 41
Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell
75
:805–816 (1993).
Hopkin, K. Yeast two-hybrid systems: more than bait and fish. J. NIH Res.
8
:27–29 (1996).
Iwabuchi, K., Li, B., Bartel, P. & Fields, S. Use of the two-hybrid system to identify the domain of p53 involved in
oligomerization. Oncogene
8
:1693–1696 (1993).
Kuo, H. J., Maslen, C. L., Keene, D. R. & Glanville, R. W. Type VI collagen anchors endothelial basement membranes by
interacting with type IV collagen. J. Biol. Chem.
272:
26522–26529 (1997).
Li, B. & Fields, S. Identification of mutations in p53 that affect its binding to SV40 T antigen by using the yeast two-hybrid
system. FASEB J.
7
:957–963 (1993).
Luban, J. & Goff, S. P. The yeast two-hybrid system for studying protein-protein interactions. Current Opinion in
Biotechnol.
6
:59–64 (1995).
McNabb, D. S. & Guarente, L. Genetic and biochemical probes for protein-protein interactions. Curr. Opin. Biotechnol.
7
(5):554–559 (1996).
Mendelsohn, A. R. & Brent, R. Biotechnology applications of interaction traps/two-hybrid systems. Curr. Opinion in
Biotechnol.
5
:482–486 (1994).
Pringle, J. R., Roach, J. R. & Jones, E. W., eds. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell
Cycle and Cell Biology (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY) (1997).
Ruden, D. M. Activating regions of yeast transcription factors must have both acidic and hydrophobic amino acids.
Chromosoma
101
:342–348 (1992).
Ruden, D. M., Ma, J., Li, Y., Wood, K. & Ptashne, M. Generating yeast transcriptional activators containing no yeast protein
sequences. Nature
350
:250–251 (1991).
Schultz, J., Ferguson, B. & Sprague, Jr., G. F. Signal transduction and growth control in yeast. Curr. Opinion in Genet.
Devel.
5
:31–37 (1995).
Serebriiskii, I., Estojak J., Berman M. & Golemis E. A. Approaches to detecting false positives in yeast two-hybrid systems.
Biotechniques
28
:328–336 (2000).
van Aelst, L., Barr, M., Marcus, S., Polverino, A. & Wigler, M. Complex formation between RAS and RAF and other
protein kinases. Proc. Natl. Acad. Sci. USA 90:6213–6217 (1993).
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 32 of 41
Appendix A: Mate & Plate Library Construction
A. Host Strain Information
The phenotypes and complete genotypes of Y2HGold and Y187 (the library strain) are shown in Tables II
and III. For additional information on the growth and maintenance of yeast, see the supporting
Matchmaker protocols at www.takarabio.com. We also recommend the Guide to Yeast Genetics and
Molecular Biology (Guthrie & Fink, 1991).
Table II. Yeast Host Strain Genotypes
Strain
Genotype
a
Reporters
Transformation
Markers
Reference
Y2HGold
b, d
MATa, trp1-901, leu2-3, 112,
ura3-52, his3-200, gal4Δ, gal80Δ,
LYS2 : : GAL1
UAS
–Gal1
TATA
–His3,
GAL2
UAS
–Gal2
TATA
–Ade2
URA3 : : MEL1
UAS
–Mel1
TATA
AUR1-C MEL1
AbA
r
,
HIS3,
ADE2, MEL1
trp1, leu2
Nguyen,
unpublished
Y187
c
MATα, ura3-52, his3-200,
ade2-101, trp1-901, leu2-3, 112,
gal4Δ, gal80Δ, met–,
URA3 : : GAL1
UAS
–Gal1
TATA
–LacZ,
MEL1
MEL1, LacZ
trp1, leu2
Harper
et al. 1993
a
The GAL1, GAL2, and MEL1 upstream activating sequences (UASs) are recognized and bound by the Gal4 BD. The trp1, his3,
gal4, and gal80 mutations are all deletions; leu2-3, 112 is a double mutation.
b
Y2HGold is a derivative of strain PJ69-2A (James et al. 1996). The ade2-101 mutation in the precursor strain, PJ69-2A, was
replaced (by recombination) with the GAL2–Ade2 reporter construct. In the absence of GAL4, Y2HGold displays the Ade–
phenotype.
c
The LacZ reporter construct was integrated into the yeast genome by homologous recombination at the ura3-52 mutation (A. Holtz,
unpublished). Recombinants were selected on SD/–Ura. The met– phenotype in this strain is unstable.
d
The AUR1-C reporter construct was integrated into the yeast genome by homologous recombination at the ura3-52 mutation (Y.
Nguyen, unpublished). Recombinants were selected on SD/–Ura. The met– phenotype in this strain is unstable.
Table III. Phenotype Testing on Various SD Media
Strain
SD/–Ade
SD/–His
SD/–Leu
SD/–Trp
SD/–Ura
Y2HGold
–
–
–
–
+
Y187
–
–
–
–
+
Y2HGold
[pGBKT7-53]
–
–
–
+
+
Y187 [pGADT7-
T]
–
–
+
–
+
Control Diploid
1
+
+
+
+
+
1
Diploid strain derived from mating Y2HGold [pGBKT7-53] with Y187 [pGADT7-T].
B. General Considerations for Mate & Plate Library Construction
Matchmaker Libraries may be cloned into one of several commonly used Gal4 AD vectors. Three of these
are described below. To find out which vector your library was constructed in, refer to the Certificate of
Analysis (CofA) included with your library. Vector maps are available at www.takarabio.com.
• Libraries constructed in pGADT7-Rec
Mate & Plate Libraries cloned in pGADT7-Rec are constructed using our Make Your Own “Mate &
Plate” Library System (Cat. No. 630490). pGADT7-Rec offers a simple and efficient method for
constructing two-hybrid libraries via recombination-mediated cloning in vivo directly in S. cerevisiae. See
the Make Your Own “Mate & Plate” Library System User Manual (PT4085-1) at www.takarabio.com
for a description of the procedure.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 33 of 41
• Libraries constructed in pGADT7-RecAB
Normalized Mate & Plate libraries are constructed in pGADT7-RecAB. cDNA synthesized using our
SMART® technology is normalized to reduce the proportion of highly abundant transcripts. Normalized
cDNA is SfiI-digested, and cloned into the SfiI A/B sites of pGADT7-RecAB.
Once cloned, the library is amplified in E. coli, rescued, and used to transform yeast strain Y187. The
resulting colonies are harvested at high density in freezing medium and immediately aliquoted and frozen
at –70°C.
• Libraries Constructed in pACT2
Mate & Plate Libraries that are cloned in pACT2 were prepared using a modified Gubler & Hoffman
procedure (1983). Once cloned, the library is amplified in E. coli. The library plasmid is then isolated and
used to transform yeast strain Y187. The resulting colonies are harvested, aliquoted, and frozen at –70°C.
C. Library Priming
Matchmaker cDNA libraries are oligo(dT)-primed, or oligo(dT) + random-primed, as stated on the
Certificate of Analysis (CofA); please refer to your CofA for the specific type of oligo(dT) primer used in
the first-strand synthesis. The “lock-docking” oligo(dT)25d(A/C/G) primer contains a degenerate
nucleotide site that positions the primer at the junction of the poly-A tail and the transcript proper (Borson
et al. 1992). This primer eliminates synthesis of lengthy poly(dT) regions and thereby enhances the
representation of full-length clones and 3' ends in the library (Borson et al. 1992). Random priming may
lead to a greater representation of all portions of the gene, including amino-terminal and internal domains,
regardless of mRNA secondary structure; random priming also generates a wider size-range of cDNA.
For oligo(dT) + random-primed libraries, separate first-strand syntheses are performed with each type of
primer; after second-strand synthesis (before ligation to the adaptor), the cDNAs are pooled in roughly
equal proportions.
Unidirectional libraries are made with oligo(dT) primers that have one vector-compatible restriction
enzyme site. The other site is added (with sticky ends) by the adaptor that is ligated to the cDNA. Thus,
digestion with one restriction enzyme ensures the cDNA’s proper orientation when ligated to a vector that
has been digested with the appropriate two enzymes.
D. Adaptors and Linkers
Please refer to the Product Certificate of Analysis (CofA) for information on the adaptor or linker used in
the construction of your Matchmaker Library. pACT2-based libraries are made using adaptor ligation.
pGADT7-Rec- and pGADT7-RecAB-based libraries are constructed using Takara Bio’s SMART
technology, as outlined in the user manual of the ”Make Your Own Mate & Plate Library System” (Cat.
No. 630490)
NOTES:
• The open reading frame of the insert starts at the codon immediately following the C-terminal codon
(amino acid 881) of the GAL4 AD, not within the adaptor.
• If an EcoRI linker is used, the cDNA is methylated to protect any internal EcoRI sites.
• If an adaptor is used in the construction of nondirectionally cloned libraries, no methylation or
restriction enzyme digestion of the cDNA is required; therefore, any internal EcoRI sites present in
the cDNA will not be cut.
• If an adaptor is used in the construction of unidirectionally cloned libraries, the cDNA is methylated
to protect the alternative site.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 34 of 41
• If the library is synthesized using EcoRI/NotI/SalI adaptors, you may excise the inserts from the
vector using sites within the adaptor.
• For information about pGADT7-Rec, please refer to Vector Information Packet PT3530-5, supplied
with all libraries constructed in this vector.
E. cDNA Size Fractionation
All ds cDNA is size-fractionated to remove unincorporated primers, unligated adaptors, and adaptor
dimers; this process also removes low-molecular-weight (i.e., <400 bp) incomplete cDNAs. Matchmaker
Libraries have a wide range of insert sizes (generally >400 bp), which may be an advantage in a two-
hybrid library screening (Fritz & Green, 1992; Fields & Sternglanz, 1994). See the CofA for quality
control information about insert size.
F. Normalized Mate & Plate Libraries
Please refer to the CofA for information on the normalization procedure used in the construction of the
Normalized Libraries.
G. Insert Size Range and Average Insert Size
Sizes are determined by running the cDNA on a gel prior to cloning, and comparing the profile to MW
size markers.
H. Library Amplification
Unless otherwise stated on the CofA, libraries constructed in pACT2 and pGADT7-RecAB were
amplified once in E. coli. Libraries constructed in pGADT7-Rec are not amplified since they are made
directly in yeast.
I. Quality Control of the Original cDNA Library
• cDNA size range and average cDNA size are determined by running the cDNA on a gel prior to
cloning and comparing the profile to MW size markers. The average insert size range is 0.5 kb to
4.0 kb.
• Number of independent clones is estimated before amplification. All libraries are guaranteed to
have at least 1 x 10
6
independent clones and are representative of the mRNA population
complexity. See Ausubel et al. (1994) for a general discussion of library size.
J. Quality Control for the Mate & Plate Libraries
Refer to the CofA included with your library.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 35 of 41
Appendix B: Library Titering
A. General Considerations
• Diluted libraries are always less stable than undiluted libraries. Therefore, once the library
dilutions are made, plate them within the next hour, before misleading reductions in titer can
occur.
• Use proper sterile technique when aliquoting and handling libraries.
• Design and use appropriate controls to test for cross-contamination.
• Always use the recommended concentration of antibiotic (i.e., kanamycin) in the medium to
suppress growth of contaminating bacteria.
B. Library Titering
1. Materials:
• YPDA Broth (Appendix D)
• SD/–Leu (100-mm plates) (Appendix D)
• Sterile glass spreading rod, bent Pasteur pipette, or 5-mm glass beads for spreading
culture on plates.
2. Transfer the 10-µl library aliquot (reserved from Section VIII) to 1 ml of YPDA Broth in a 1.5-ml
microcentrifuge tube. Mix by gentle vortexing. This is Dilution A (dilution factor = 10
–2
).
NOTE: Matchmaker Gold Libraries are extremely viscous, so pay special attention during
aliquoting.
3. Remove 10 µl from Dilution A and add it to 1 ml of YPDA Broth in a 1.5-ml microcentrifuge
tube. Mix by gentle vortexing. This is Dilution B (dilution factor = 10
–4
).
4. Add 10 µl from Dilution A to 50 µl of YPDA Broth in a 1.5-ml microcentrifuge tube. Mix by
gentle vortexing. Spread the entire mixture onto an SD/–Leu plate.
5. Remove 50 µl aliquots from Dilution B and spread onto separate SD/–Leu plates as above.
6. Invert the plates and incubate at 30°C for 3–5 days.
NOTE: Colony size will vary, depending on the insert.
7. Count the number of colonies on plates having 30–300 colonies.
8. Calculate the titer (cfu/ml) as follows:
Number of colonies = cfu/ml
plating volume (ml) x dilution factor
NOTE: Due to slight variability in pipettes and pipetting techniques, a 2–5-fold range in titer
calculations is not unusual.
Example calculation
• No. of colonies on plate = 100
• Plating volume = 0.05 ml
• Dilution factor = 10
–4
100 = 2 x 10
7
cfu/ml
0.05 ml x 10
-4
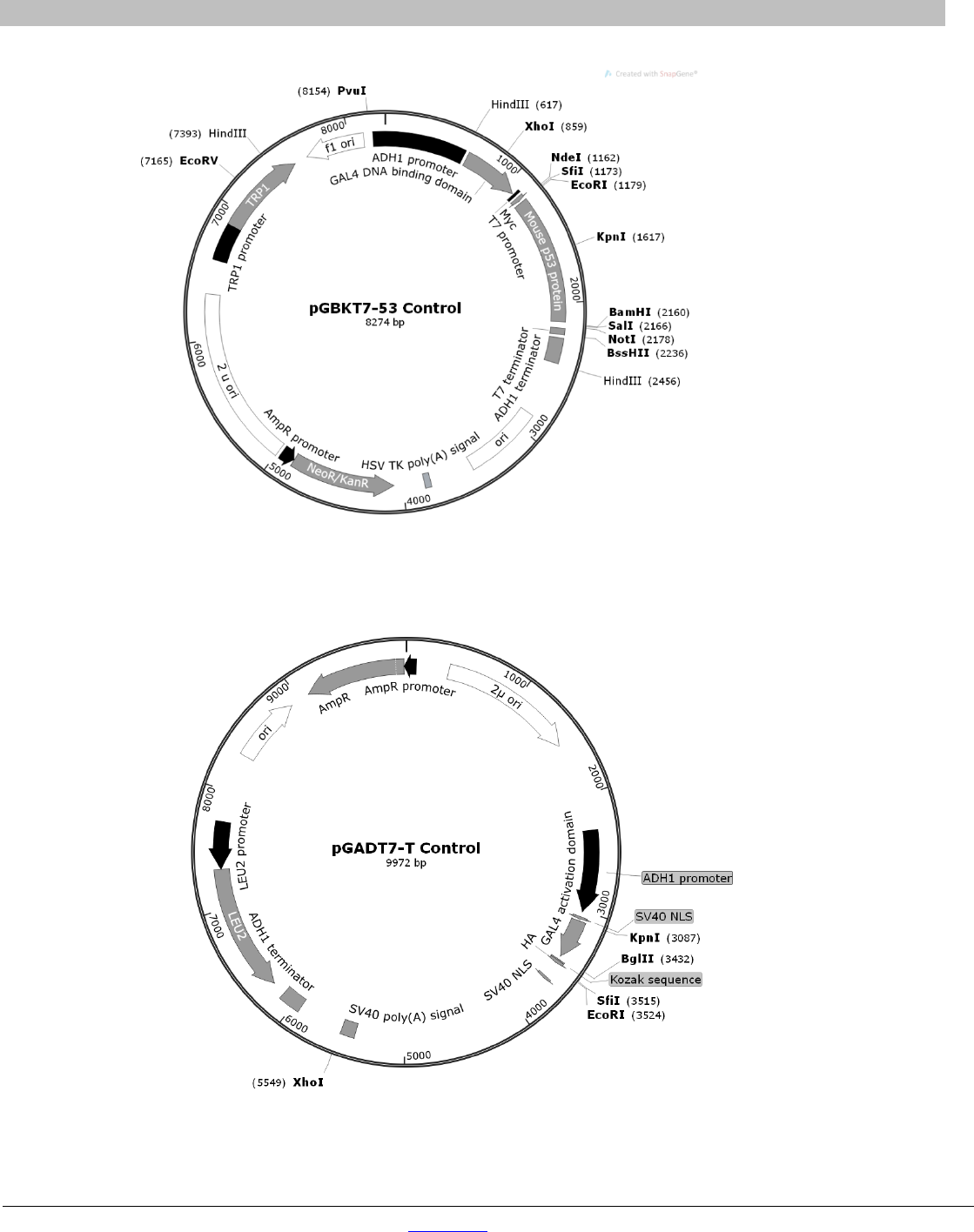
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 36 of 41
Figure 10. Map of pGBKT7-53 DNA-BD control plasmid. pGBKT7-53 is a positive control plasmid that encodes a fusion of
the murine p53 protein (a.a. 72–390) and the GAL4 DNA-BD (a.a. 1–147). The murine p53 cDNA (GenBank Accession No.
K01700) was cloned into pGBKT7 at the EcoRI and BamHI sites. The p53 insert was derived from the plasmid described in
Iwabuchi et al. (1993); plasmid modification was performed at Takara Bio. pGBKT7-53 has not been sequenced.
Figure 11. Map of pGADT7-T AD control plasmid. pGADT7-T is a positive control plasmid that encodes a fusion of the SV40
large T antigen (a.a. 87–708) and the GAL4 AD (a.a. 768–881). The SV40 large T antigen cDNA (GenBank Locus SV4CG),
which was derived from the plasmid referenced in Li & Fields (1993), was cloned into pGADT7; plasmid modification was
performed at Takara Bio. pGADT7-T has not been sequenced and it is not known whether any of the sites are unique.
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 37 of 41
Appendix D: Yeast Growth Media & Supplements
A. Ready-to-go Media Pouches Available from Takara Bio
Takara Bio offers media sets with a complete assortment of mixes in convenient, “ready-mixed” foil
pouches, for use with the Matchmaker Gold Yeast Two-Hybrid System. We also provide the media
supplements Aureobasidin A and X-alpha-Gal in an optional combination with the media set. In addition,
each of the media mixes as well as these two supplements may be purchased separately.
• See Table IV for a list of the components of the Yeast Media Set 2 (Cat. No. 630494) and the
Yeast Media Set 2 Plus (Cat. No. 630495). The Yeast Media Set 2 Plus also contains the
additional media supplements Aureobasidin A and X-alpha-Gal, which are required for the
protocols described in this user manual.
• See Table V for information for purchasing each of the media mixes separately, in packs of 10
pouches.
• See Table VI for information on preparing all additional required media supplements and
purchasing Aureobasidin A and X-alpha-Gal separately.
Table IV. Components of Yeast Media Set 2 & Yeast Media Set 2 Plus
Media Pouch
Quantity of Pouches Supplied
Volume of Media Each Pouch Makes
YPDA Broth
2
0.5 L
YPDA with Agar
1
0.5 L
SD/–Leu Broth
1
0.5 L
SD/–Leu with Agar
1
0.5 L
SD/–Trp Broth
1
0.5 L
SD/–Trp with Agar
1
0.5 L
SD/–Leu/–Trp with Agar
10
0.5 L
SD/–Ade/–His/–Leu/–Trp with Agar
1
0.5 L
Additional Components in Yeast Media Set 2 Plus
X-alpha-Gal
250 mg
—
Aureobasidin A
1 mg
—
Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 38 of 41
Table V. Individual Yeast Media Pouches for Matchmaker Gold Protocols
Yeast Media Pouches
Takara Bio
Cat. No.
Quantity of
Pouches
Supplied
Volume of Media
Each Pouch
Makes
Rich Media (for Routine Culturing of Untransformed Yeast)
YPDA Broth
YPDA with Agar
630306
630307
10
10
0.5 L
0.5 L
Minimal Media Single Dropouts (SDO)
SD–Trp Broth
SD–Trp with Agar
SD–Leu Broth
SD–Leu with Agar
630308
630309
630310
630311
10
10
10
10
0.5 L
0.5 L
0.5 L
0.5 L
Minimal Media Double Dropouts (DDO)
SD–Leu/–Trp Broth
SD–Leu/–Trp with Agar
630316
630317
10
10
0.5 L
0.5 L
Minimal Media Triple Dropouts (TDO)
SD–His/–Leu/–Trp Broth
SD–His/–Leu/–Trp with Agar
630318
630319
10
10
0.5 L
0.5 L
Minimal Media Quadruple Dropouts (QDO)
SD–Ade/–His/–Leu/–Trp Broth
SD–Ade/–His/–Leu/–Trp with Agar
630322
630323
10
10
0.5 L
0.5 L
Table VI. Additional Media & Media Supplements Required for a Two-Hybrid Screen
Freezing Medium
Preparation Instructions
YPDA Medium & 25% glycerol
See Section E of Appendix D
Supplement Name
Takara Bio Cat. No.
1
Stock Solution Concentration
Aureobasidin A
630466
630499
500 µg/ml (see Section F of Appendix D)
X-alpha-Gal (250 mg)
630462
630463
20 mg/ml in dimethyl formamide
Kanamycin sulfate
—
50 mg/ml stock solution
Dimethyl formamide
—
—
1
Unless otherwise specified.
B. General Media Preparation Instructions
• Prepare media by dissolving pouch contents in 500 ml ddH
2
O, autoclave for 15 min at 121˚C, and
allow to cool before use (or filter-sterilize broth media). Do not over-autoclave.
• This media does not usually require pH adjustment, but if your source water is particularly acidic,
you may need to adjust the pH of the media to 5.8.
• For additional information on preparing media from the pouches, please see the Takara Bio Yeast
Media Protocol-at-a-Glance (PT4057-2) at www.takarabio.com
C. 2X YPDA Broth
Reconstitute one YPDA Broth pouch in 250 ml ddH
2
O and sterilize.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 39 of 41
D. 0.5X YPDA Broth
Reconstitute one YPDA Broth pouch in 1 L ddH
2
O and sterilize.
E. Freezing Medium
Mix 100 ml YPDA (sterile) and 50 ml 75% glycerol (sterile).
F. Aureobasidin A Stock Solution Recipe:
Dissolve 1 mg Aureobasidin A (Cat. No. 630466) in 2 ml of absolute ethanol for a stock concentration of
500 µg/ml. Store at 4°C.
G. Aureobasidin A Working Concentration
Add just 200 µl of the Aureobasidin A stock solution to 500 ml of dropout agar media for yeast two-
hybrid screening, yielding a final concentration of 200 ng/ml.
H. X-alpha-Gal Stock Solution Recipe
Dissolve X-alpha-Gal (Cat. No. 630463) at 20 mg/ml in dimethylformamide (DMF). Store X-alpha-Gal
solutions at –20°C in the dark.
I. X-alpha-Gal Working Concentration
Add 1 ml of the X-alpha-Gal stock solution to 500 ml dropout media for yeast two-hybrid screening.
J. SDO Agar Plates
Single dropout (–Trp) or (–Leu) media is prepared as follows:
• Prepare 500 ml agar media using SD/–Trp (or SD/–Leu) with Agar, autoclave, and cool to
55–60°C in a water bath.
• Mix, pour immediately, and allow to dry.
K. SDO/X Agar Plates
Single dropout (–Trp) media containing 40 µg/ml X-alpha-Gal is prepared as follows:
• Prepare 500 ml agar media using SD/–Trp with Agar, autoclave, and cool to 55–60°C in a water
bath.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Mix, pour immediately, and allow to dry.
L. SDO/X/A Agar Plates
Single dropout (–Trp) media containing 40 µg/ml X-alpha-Gal and 200 ng/ml Aureobasidin A is prepared
as follows:
• Prepare 500 ml agar media using SD/–Trp with Agar, autoclave, and cool to 55–60°C in a water
bath.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Add 200 µl of Aureobasidin A stock solution.
• Mix, pour immediately, and allow to dry.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 40 of 41
M. DDO Agar Plates
Double dropout media is prepared as follows:
• Prepare 500 ml agar media using SD–Leu/–Trp with Agar, autoclave, and cool to 55–60°C in a
water bath.
• Mix, pour immediately, and allow to dry.
N. DDO/X Agar Plates
Double dropout media containing 40 µg/ml X-alpha-Gal is prepared as follows:
• Prepare 500 ml agar media using SD–Leu/–Trp with Agar, autoclave, and cool to 55–60°C in a
water bath.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Mix, pour immediately, and allow to dry.
O. DDO/X/A Agar Plates
Double dropout media containing 40 µg/ml X-alpha-Gal and 200 ng/ml Aureobasidin A is prepared as
follows:
• Prepare 500 ml agar media using SD–Leu/–Trp with Agar, autoclave, and cool to 55–60°C in a
water bath.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Add 200 µl of Aureobasidin A stock solution.
• Mix, pour immediately, and allow to dry.
P. QDO/X Agar Plates
Quadruple dropout media containing 40 µg/ml X-alpha-Gal is prepared as follows:
• Prepare 500 ml agar media using SD/–Ade/–His/–Leu/–Trp with Agar, autoclave, and cool to 55–
60°C in a waterbath.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Mix, pour immediately, and allow to dry.
Q. QDO/X/A Agar Plates
Quadruple dropout media containing 40 µg/ml X-alpha-Gal and 200 ng/ml Aureobasidin A is prepared as
follows:
• Prepare 500 ml agar media using SD/–Ade/–His/–Leu/–Trp with Agar, autoclave, and cool to 55–
60°C in a waterbath.
• Add 200 µl of Aureobasidin A stock solution.
• Add 1 ml of X-alpha-Gal Stock Solution.
• Mix, pour immediately, and allow to dry.
R. Kanamycin Supplement to Yeast Media
Kanamycin sulfate can be added to all yeast media at a final concentration of 50 µg/ml to stop bacterial
contamination. Please note that kanamycin does not stop contaminating fungal growth, so proper sterile
technique must still be used. Also note that this antibiotic does not select for any plasmids in yeast.

Matchmaker Gold Yeast Two-Hybrid System User Manual
(042424)
takarabio.com
Takara Bio USA, Inc.
Page 41 of 41
Contact Us
Customer Service/Ordering
Technical Support
tel: 800.662.2566 (toll-free)
tel: 800.662.2566 (toll-free)
fax: 800.424.1350 (toll-free)
fax: 800.424.1350 (toll-free)
web: takarabio.com
web: takarabio.com
e-mail: ordersUS@takarabio.com
e-mail: technical_support@takarabio.com
Notice to Purchaser
Our products are to be used for research purposes only. They may not be used for any other purpose, including, but not limited to, use in drugs, in vitro diagnostic
purposes, therapeutics, or in humans. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to
provide a service to third parties without prior written approval of Takara Bio USA, Inc.
Your use of this product is also subject to compliance with any applicable licensing requirements described on the product’s web page at takarabio.com. It is your
responsibility to review, understand and adhere to any restrictions imposed by such statements.
©2024 Takara Bio Inc. All Rights Reserved.
All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be
registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.
This document has been reviewed and approved by the Quality Department.
