Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
1
ACCELERATED EMERGENCY USE AUTHORIZATION (EUA) SUMMARY
INFORM DIAGNOSTICS SARS-COV-2 RT-PCR ASSAY
(INFORM DIAGNOSTICS, INC.)
For In vitro Diagnostic Use
Rx Only
For use under Emergency Use Authorization (EUA) only
(The Inform Diagnostics SARS-CoV-2 RT-PCR Assay will be performed at Inform
Diagnostics, Inc. certified under the Clinical Laboratory Improvement Amendments
of 1988(CLIA), 42 U.S.C. §263a, as per the Standard Operating Procedure that was
reviewed by the FDA under this EUA.)
INTENDED USE
The Inform Diagnostics SARS-CoV-2 RT-PCR Assay is a real-time reverse transcription
polymerase chain reaction (RT-PCR) test intended for the qualitative detection of nucleic
acid from SARS-CoV-2 in nasopharyngeal, oropharyngeal, anterior nasal, and mid-
turbinate nasal swabs, as well as nasopharyngeal wash/aspirate or nasal aspirates, and
bronchoalveolar lavage (BAL) specimens from individuals suspected of COVID-19 by
their healthcare provider.
Testing is limited to Inform Diagnostics, Inc. located in Phoenix, AZ which is certified
under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C.
§263a and meets requirements to perform high-complexity tests.
Results are for the detection and identification of SARS-CoV-2 RNA. The SARS-CoV-2
RNA is generally detectable in respiratory specimens during the acute phase of infection.
Positive results are indicative of the presence of SARS-CoV-2 RNA; clinical correlation
with patient history and other diagnostic information is necessary to determine patient
infection status. Positive results do not rule out bacterial infection or co-infection with
other viruses. The agent detected may not be the definite cause of disease. Laboratories
within the United States and its territories are required to report all positive results to the
appropriate public health authorities.
Negative results do not preclude SARS-CoV-2 infection and should not be used as the
sole basis for patient treatment or other patient management decisions. Negative results
must be combined with clinical observations, patient history, and epidemiological
information.
Testing with the Inform Diagnostics SARS-CoV-2 RT-PCR Assay is intended for use by
qualified and trained laboratory personnel specifically instructed and trained in the
techniques of real-time PCR and in vitro diagnostic procedures. The Inform Diagnostics
SARS-CoV-2 RT-PCR Assay is only for use under the Food and Drug Administration’s
Emergency Use Authorization.
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
2
DEVICE DESCRIPTION AND TEST PRINCIPLE
The Inform Diagnostics SARS-CoV-2 RT-PCR Assay is a real-time reverse transcription
polymerase chain reaction test. The assay uses primers and probes that were developed
and validated under the Emergency Use Authorization (EUA) for the CDC 2019-nCoV
Real-Time RT-PCR Diagnostic Panel and are designed to detect RNA from SARS-CoV-
2 in respiratory specimens from patients suspected of COVID-19 by their healthcare
provider. The test uses two primer and probe sets to detect two regions in the SARS-
CoV-2 nucleocapsid (N) gene (N1 and N2), and one primer and probe set to detect
human RNase P (RP) in control samples and clinical specimens. Three separate master
mixes for each target are prepared and run with the Inform Diagnostics Assay.
RNA is isolated from respiratory specimens including nasopharyngeal, oropharyngeal,
anterior nasal, and mid-turbinate nasal swabs as well as nasopharyngeal wash/aspirate or
nasal aspirates and BAL specimens using the ViralXpress DNA/RNA Extraction Reagent
(Millipore, Cat # 3095). Nucleic acid is manually extracted from 50 µL of acceptable
specimen with the addition of carrier RNA into the lysis buffer. RNA is reverse
transcribed to cDNA and subsequently amplified using either the Applied Biosystems
7500 Fast Dx Real-Time PCR Instrument with software version 1.4 or the QuantStudio
with software version 1.0.3. During the amplification process, the probe anneals to a
specific target sequence located between the forward and reverse primers. During the
extension phase of the PCR cycle, the 5′ nuclease activity of Taq polymerase degrades
the bound probe, causing the reporter dye (FAM) to separate from the quencher dye
(BHQ-1), generating a fluorescent signal. Fluorescence intensity is monitored at each
PCR cycle.
INSTRUMENTS USED WITH TEST
The Inform Diagnostics SARS-CoV-2 Assay is to be used with the following Applied
Biosystems PCR platforms:
• 7500 Fast Dx Real-Time PCR Instrument (ABI7500) with software version 1.4
• QuantStudio with software version 1.0.3
The QIAgility (Qiagen, Cat # Q080957) with software version 4.17.1 is used to prepare
and dispense the RT-PCR master mix into the reaction plate.
REAGENTS AND MATERIALS
Reagent Manufacturer and Description
Catalog #
Manufacturer
ViralXpress DNA/RNA Extraction Reagent
3095
Millipore
Carrier RNA
1017647
Qiagen
TaqPath 1-Step RT-qPCR Mater Mix, CG
A15300, A15299
ThermoFisher Scientific
COVID-19_N1-F Primer (forward primer)
10006606
Integrated DNA Technologies
COVID-19_N1-R Primer (reverse primer)
10006606
Integrated DNA Technologies
COVID-19_N1-P Probe (N1 probe)
10006606
Integrated DNA Technologies
COVID-19_N2-F Primer (forward primer)
10006606
Integrated DNA Technologies
COVID-19_N2-R Primer (reverse primer)
10006606
Integrated DNA Technologies
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
3
Reagent Manufacturer and Description
Catalog #
Manufacturer
COVID-19_N2-P Probe (N2 probe)
10006606
Integrated DNA Technologies
RP-F Primer (forward primer)
10006606
Integrated DNA Technologies
RP-R Primer (reverse primer)
10006606
Integrated DNA Technologies
RP-P Probe (RNase P probe)
10006606
Integrated DNA Technologies
Hs_RPP30 Positive Control
10006626
Integrated DNA Technologies
2019-nCoV_N_Positive Control
10006625
Integrated DNA Technologies
MicroAmp Optical 96-Well Reaction plate
4481192
Life Technologies
MicroAmp Optical Adhesive Film
4311971
Life Technologies
CONTROLS TO BE USED WITH THE COVID-19 RT-PCR
1) A no template control (NTC) is needed to check for contamination of RT-PCR
assay reagents. Molecular grade, nuclease-free water is used in place of sample
nucleic acid for this control. The NTC is used on every assay plate.
2) The positive control is prepared using the 2019-nCoV_N_Positive Control (IDT,
Cat # 10006625) and Hs_RPP30 Positive Control (IDT, Cat # 10006626).
Positive template control is needed to verify PCR reagent integrity as well as
proper assay set-up of the RT-PCR reactions for the N1, N2, and RNase P genes.
The positive control is used on every assay plate starting at master mix addition at
a final N1, N2, and RNase P template concentration of 50 copies/µL. The 2019-
nCoV_N_Positive Control is commercially supplied from IDT and is made of in
vitro transcribed and purified plasmid DNA targets that contains one copy each of
N1 and N2. The Hs_RPP30 Positive Control is RNase P template supplied from
IDT that Inform Diagnostics also incorporates into their SARS-CoV-2 positive
control.
3) A negative extraction (NEC) control is a nasopharyngeal swab sample from a
previously confirmed SARS-CoV-2 negative patient. This control is used as a
negative control as well as an extraction control to monitor for any cross-
contamination during the analytical process and to verify the success of RNA
extraction and sample integrity. A NEC is used in each extraction batch.
4) RNase P is co-extracted and amplified from all patient samples as an internal
control to assess the extraction efficiency and specimen quality. This also serves
as an extraction control to ensure that samples resulting as negative contain
nucleic acid for testing. Detection of the RNase P gene in patient test samples
verifies successful extraction of the sample, proper assay setup, sample integrity,
and efficient sample collection. No additional RNase P is added to clinical
samples prior to performing the extraction procedure.
INTERPRETATION OF RESULTS
All test controls must be examined prior to interpretation of patient results. If the
controls are not valid, the patient results cannot be interpreted (Refer to Table 1 for a
summary of control results).

Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
4
1) COVID-19 RT-PCR Test Controls – Positive, Extraction, Internal, and NTC:
• No template controls should be negative (Ct Not Detected) for all targets. If the
N1, N2, or RNase P targets exhibit positive fluorescence above the threshold (Ct
≤ 40), it is possible that contamination occurred, or that the assay was setup
improperly. The RT-PCR run is invalid. The user is instructed to repeat the RT-
PCR using residual extracted material. If the repeat NTC result is positive for any
of the assay targets, re-extract residual clinical specimens and fresh controls and
re-test all samples.
• The positive control contains both the 2019-nCoV_N_Positive Control and
Hs_RPP30 and should therefore, be positive for the N1 and N2 targets (Ct ≤ 40)
as well as the RNase P target (Ct ≤ 40). Negative results with N1, N2, or RNase P
targets invalidates the run and suggests the assay may have been set up
incorrectly, the integrity of the primers/probes could have been compromised, or
potential carry-over of PCR inhibitors. The user is instructed to repeat the RT-
PCR step using residual extracted material. If the repeat test result is negative for
the N1/N2 and/or RNase P targets, re-extract residual clinical specimens and fresh
controls and repeat RT-PCR.
• The negative extraction control (negative clinical sample) should be negative for
N1, N2 (Ct Not Detected or Ct > 40), and positive for the RNase P target (Ct ≤
40). If positive results are obtained for N1 and N2 targets, contamination of
nucleic acid extraction reagents or cross-contamination of samples may have
occurred. Failure of the control to yield a RNase P Ct value of ≤ 40 may indicate
improper extraction of nucleic acid, carry-over of PCR inhibitors, or insufficient
cellular material. The extraction run and the RT-PCR run are invalid and should
be repeated using new extracted material from residual patient samples and fresh
controls.
• RNase P is co-extracted and amplified from all patient samples as an internal
control to assess the extraction efficiency and specimen quality. This also serves
as a positive extraction control to ensure that samples resulting as negative
contain nucleic acid for testing. Detection of the RP gene in patient test samples
verifies successful extraction of the sample, proper assay setup, sample integrity,
and efficient sample collection.
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
5
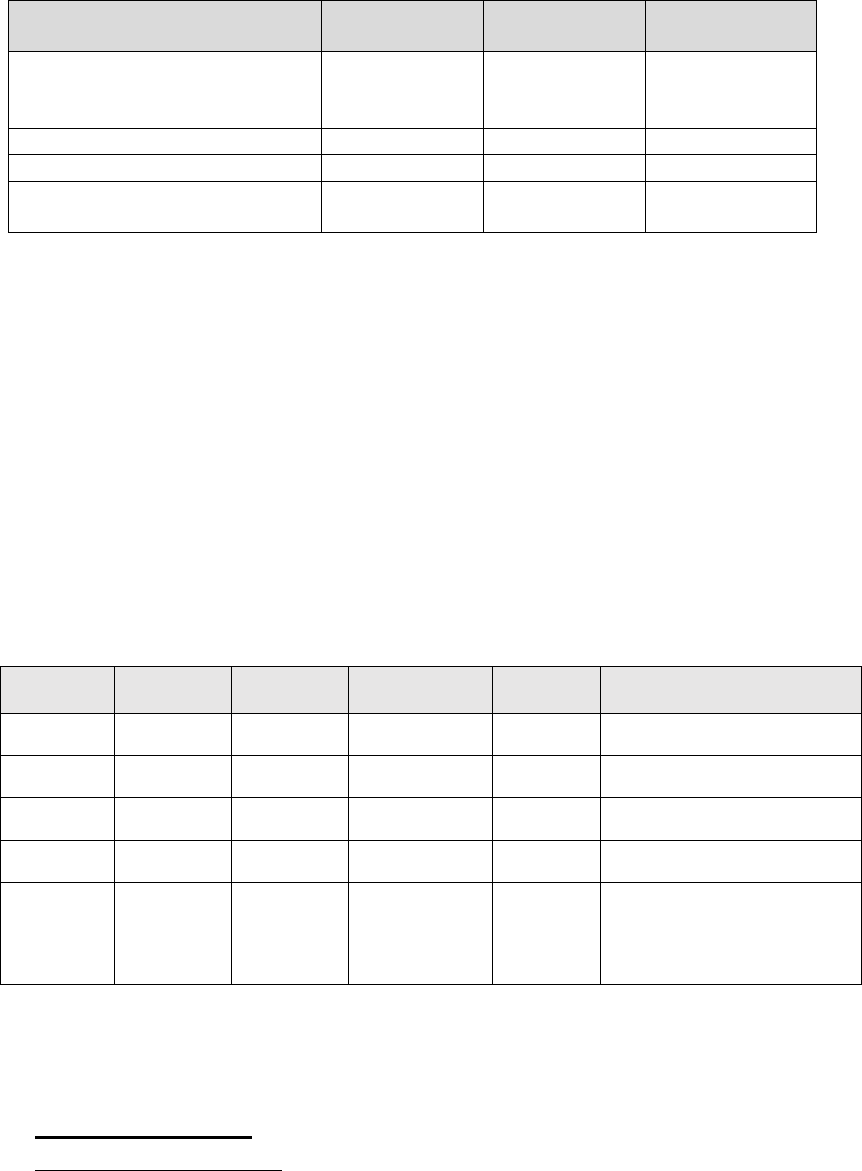
Table 1. Ct Values for Controls that Must be Observed to Obtain Valid Results
Control
Expected N1
Result
Expected N2
Result
Expected RP
Result
2019-nCoV_N_ Positive Control
with Hs_RPP30 Positive Control
(N1, N2, RP template)
Ct ≤ 40 Ct ≤ 40 Ct ≤ 40
No Template Control (NTC)
Not Detected
Not Detected
Not Detected
Negative Extraction Control
Not Detected
Not Detected
Ct ≤ 40
Internal RNase P Control Not Detected Not Detected
Ct ≤ 40 / Not
Detected*
Not Detected; no detectable signal or Ct > 40
If the results obtained with the Positive, Internal, and No Template Controls do not meet
the criteria shown, the results from the entire batch of samples are considered invalid and
repeat testing must be performed using residual extracted nucleic acid. If the negative
extraction control does not meet the acceptability criteria, all specimens in the batch
should be re-extracted from residual clinical samples and the RT-PCR assay should be re-
run.
Assessment of clinical specimen test results should be performed after the positive and
negative controls have been examined and determined to be valid and acceptable. If the
controls are not valid, the patient results cannot be interpreted. Please see the table below
(Table 2) for guidance on interpretation and reporting of results.
Table 2: Interpretation of Patient Results Using the Inform Diagnostics SARS-
CoV-2 RT-PCR Test
N1
(Ct ≤ 40)
N2
(Ct ≤ 40)
RNase P
(Ct ≤ 40)
Interpretation
Report
Result
Actions
+
+
+/-
*
SARS-CoV-2
Detected
POSITIVE
Reported to sender and appropriate
public health authorities.
+
-
+/-
*
SARS-CoV-2
Detected
POSITIVE
Reported to sender and appropriate
public health authorities.
-
+
+/-
*
SARS-CoV-2
Detected
POSITIVE
Reported to sender and appropriate
public health authorities.
-
-
+
SARS-CoV-2
Not Detected
NEGATIVE
Reported to sender and appropriate
public health authorities.
- - -
Invalid test INVALID
Repeat extraction and RT-PCR. If
the repeated result remains invalid,
consider collecting a new specimen
from the patient.
*If either N1 or N2 is positive, it is not necessary to have positive signal from the RNase P probe.
PERFORMANCE EVALUATION
1) Analytical Sensitivity:
Limit of Detection (LoD):
The LoD of the Inform Diagnostics Test was determined using synthetic SARS-CoV-
2 viral RNA from Twist Bioscience (Cat # MT007544.1). A preliminary LoD was
determined by testing serial dilutions (1000 copies/μL – 10 copies/μL) of synthetic

Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
6
RNA spiked into pooled clinical negative, nasopharyngeal swab or oropharyngeal
swab matrix using either one replicate or three replicates at each target level. Spiked
samples were tested with the Inform Diagnostics Test following extraction with the
ViralXpress Reagent on both claimed PCR instruments. Two preliminary
concentrations of 20 copies/µL and 15 copies/µL were chosen for confirmatory
testing with 20 individual extraction replicates on both the ABI7500 and QuantStudio
platforms.
The established LoD of the Inform Diagnostics SARS-CoV-2 RT-PCR Assay was 20
copies/µL. The results of the LoD confirmatory study are summarized below.
Table 3. LoD Verification Study Results
Instrument
Concentration
(copies/
µ
L)
Average Ct Values
# Detected / Total
Tested
N1
N2
N1
N2
ABI7500
20
30.21
36.22
20/20
20/20
15
34.60
37.61
8/20
8/20
QuantStudio
20
33.50
37.14
20/20
20/20
15
34.50
38.12
13/20
10/20
2) Analytical Inclusivity/Specificity:
Inclusivity:
The Inform Diagnostics SARS-CoV-2 RT-PCR Assay utilizes identical
oligonucleotide sequences for the N1 and N2 target genes to those used in the CDC
2019-Novel Coronavirus (2019-CoV) Real-Time RT-PCR Diagnostic Panel. The
inclusivity and cross-reactivity of the CDC EUA assay has been evaluated previously
and therefore, additional evaluation was not necessary. The CDC has granted a right
of reference to the performance data contained in the CDC's EUA request (FDA
submission number EUA200001) to any entity seeking an FDA EUA for a COVID-
19 diagnostic device.
Since the alignments of the CDC’s primers/probes were completed in February 2020,
an additional in silico inclusivity analysis was completed to assess the predicted
inclusivity to other deposited SARS-CoV-2 sequences in the NCBI database. In silico
testing was performed using BLASTN 2.10.1 on May 12, 2020 and confirmed 100%
nucleotide identity of the N1 and N2 primers and probes to all available SARS-CoV-
2 partial and complete genomes published by NCBI.
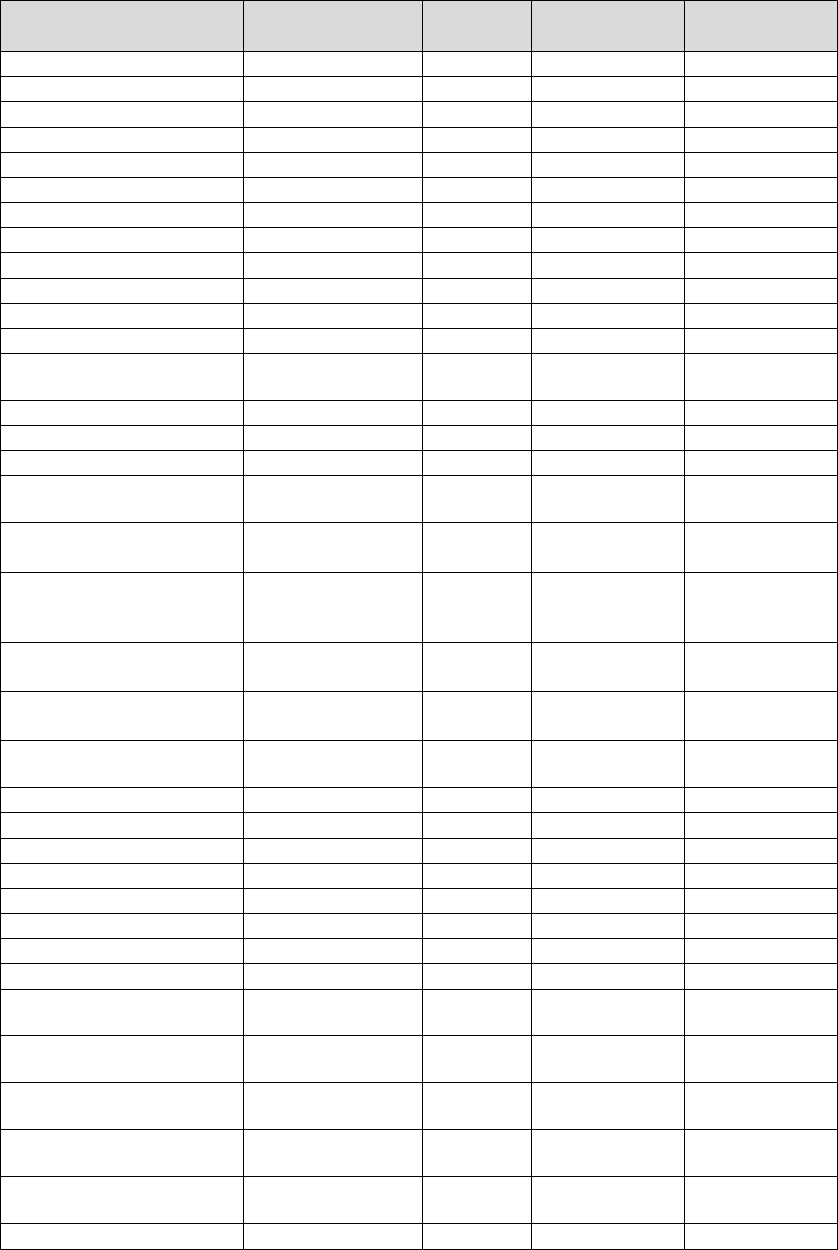
Exclusivity:
To assess for potential cross-reactivity of the Inform Diagnostics Test, an in silico
analysis of the SARS-CoV-2 N1 and N2 primer and probe sequences was performed
using BLASTN 2.10.1 against partial or complete genomes of other common
respiratory viral and bacterial pathogens listed in Table 4. None of the pathogen
sequences displayed greater than 80% homology with any of the SARS-CoV-2 N1
and N2 primers/probes.
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
7
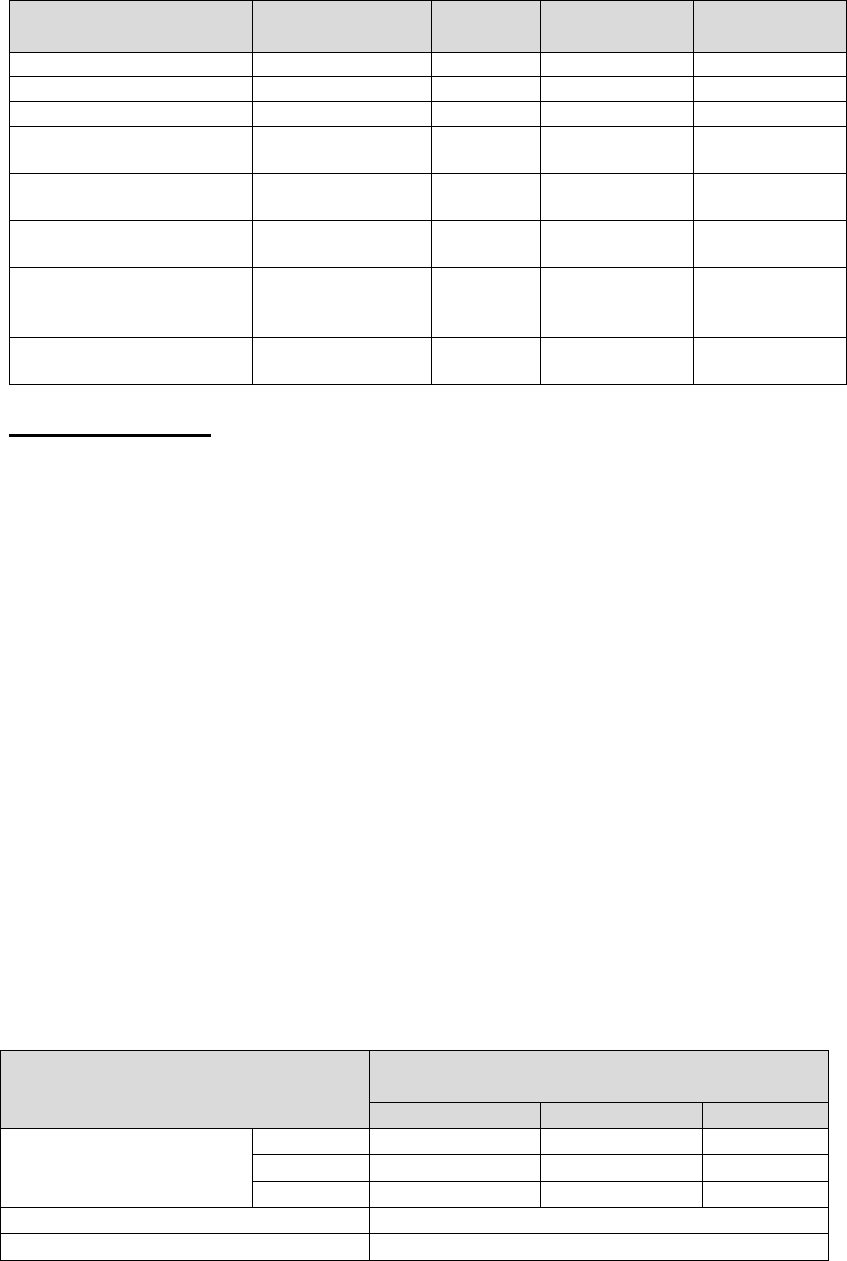
Table 4. In Silico Cross-Reactivity Analysis of N1 and N2 Oligonucleotides
Pathogen Name
GenBank
Accession ID
Genome N1 Homology N2 Homology
Human coronavirus 229E
NC_002645.1
Complete
57%
52%
Human coronavirus OC43
NC_006213.1
Complete
No homology
No homology
Human coronavirus HKU1
NC_006577.2
Complete
No homology
No homology
Human coronavirus NL63
NC_005831.2
Complete
No homology
No homology
SARS-coronavirus
NC_004718.3
Complete
57%
52%
MERS-coronavirus
NC_019843.3
Complete
No homology
No homology
Human adenovirus A
NC_001460 .1
Complete
No homology
No homology
Human adenovirus B1
NC_011203.1
Complete
No homology
No homology
Human adenovirus B2
NC_011202.1
Complete
No homology
No homology
Human adenovirus C
NC_001405 .1
Complete
No homology
No homology
Human adenovirus 54
NC_012959.1
Complete
No homology
No homology
Human adenovirus E
NC_003266.2
Complete
No homology
No homology
Human metapneumovirus
isolate 00-1
NC_039199.1
Complete No homology No homology
Parainfluenza virus 1
NC_003461.1
Complete
No homology
No homology
Parainfluenza virus 2
NC_003443.1
Complete
No homology
No homology
Parainfluenza virus 3
NC_001796.2
Complete
No homology
No homology
Parainfluenza virus 4a,
Strain: M-25
NC_021928.1
Complete No homology No homology
Influenza A virus -A/New
York/392/2004 (H3N2)
NC_007366.1-
NC_007373.1
Complete No homology No homology
Influenza A virus -
A/Puerto Rico/8/1934
(H1N1)
NC_002016.1-
NC_002023.1
Complete No homology No homology
Influenza A virus -
A/Korea/426/1968 (H2N2)
NC_007375.1-
NC_007383.1
Complete No homology No homology
Influenza B virus-
B/Lee/1940
NC_002204.1-
NC002211.1
Complete No homology No homology
Human enterovirus 68
strain Fermon
NC_038308.1
Complete No homology No homology
Human enterovirus A
NC_001612.1
Complete
No homology
No homology
Human enterovirus B
NC_001472.1
Complete
No homology
No homology
Poliovirus
NC_002058 .3
Complete
No homology
No homology
Human enterovirus D
NC_001430.1
Complete
No homology
No homology
Respiratory syncytial virus
NC_001803.1
Complete
No homology
No homology
Human rhinovirus 14
NC_001490.1
Complete
No homology
No homology
Human rhinovirus 89
NC_001617.1
Complete
No homology
No homology
Human rhinovirus C
NC_009996.1
Complete
No homology
No homology
Chlamydia pneumoniae
TW-183
NC_005043.1
Complete No homology No homology
Haemophilus influenzae
strain NCTC8143
NZ_LN831035
Partial No homology No homology
Legionella pneumophila
strain NCTC12273
NZ_LR134380.1
Partial No homology No homology
Mycobacterium
tuberculosis H37Rv
NC_000962.3
Complete No homology No homology
Streptococcus pneumoniae
strain NCTC7465
NZ_LN831051.1
Partial No homology No homology
Streptococcus pyogenes
NZ_LN831034
Partial
No homology
No homology
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
8
Pathogen Name
GenBank
Accession ID
Genome N1 Homology N2 Homology
strain NCTC8189
Bordetella pertussis 18323
NC_018518.1
Complete
No homology
No homology
Mycoplasma pneumoniae
NZ_CP010546.1
Partial
No homology
No homology
Pneumocystis jirovecii
(PJP)
MK984200
Complete No homology No homology
Candida albicans strain
L757
NC_018046
Complete No homology No homology
Pseudomonas aeruginosa
PAO1
NC_002516.2
Complete No homology No homology
Staphylococcus
epidermidis strain
ATCC14990
NZ_CP035288.1-
NZ_CP035290.1
Partial No homology No homology
Streptococcus salivarius
strain NCTC8618
NZ_LR134274.1
Partial No homology No homology
3) Clinical Evaluation:
Performance of the Inform Diagnostics SARS-CoV-2 RT-PCR Assay was evaluated
using clinical nasopharyngeal and oropharyngeal positive and negative swab
specimens that were previously tested with the Inform Diagnostics’ in-house
unmodified CDC assay. A total of 31 confirmed negative patient samples and 33
confirmed positive patient samples were tested with the unmodified CDC assay. In
addition, based on the reporting strategy for the original CDC EUA authorization, one
result was indeterminate on both PCR platforms which was excluded from the
performance analysis. Therefore, a total of 64 clinical specimens were used to assess
the clinical performance of the Inform Diagnostics SARS-CoV-2 RT-PCR Assay
(modified CDC assay) using both claimed PCR instruments including the ABI7500
Fast System and the QuantStudio.
For the 31 negative clinical NP and OP swab samples, the negative percent agreement
(NPA) between the Inform Diagnostics’ assay and the unmodified in-house CDC
EUA assay used as the comparator was 100%. For the 33 clinical positive samples
that were evaluated, 31/33 tested positive (93.9% PPA) using the Inform Diagnostics
assay when run on the ABI7500 Fast System and 32/33 tested positive (96.9%) with
the Inform Diagnostics assay when run on the QuantStudio. Qualitative results of the
clinical evaluation are shown in Tables 5 and 6.
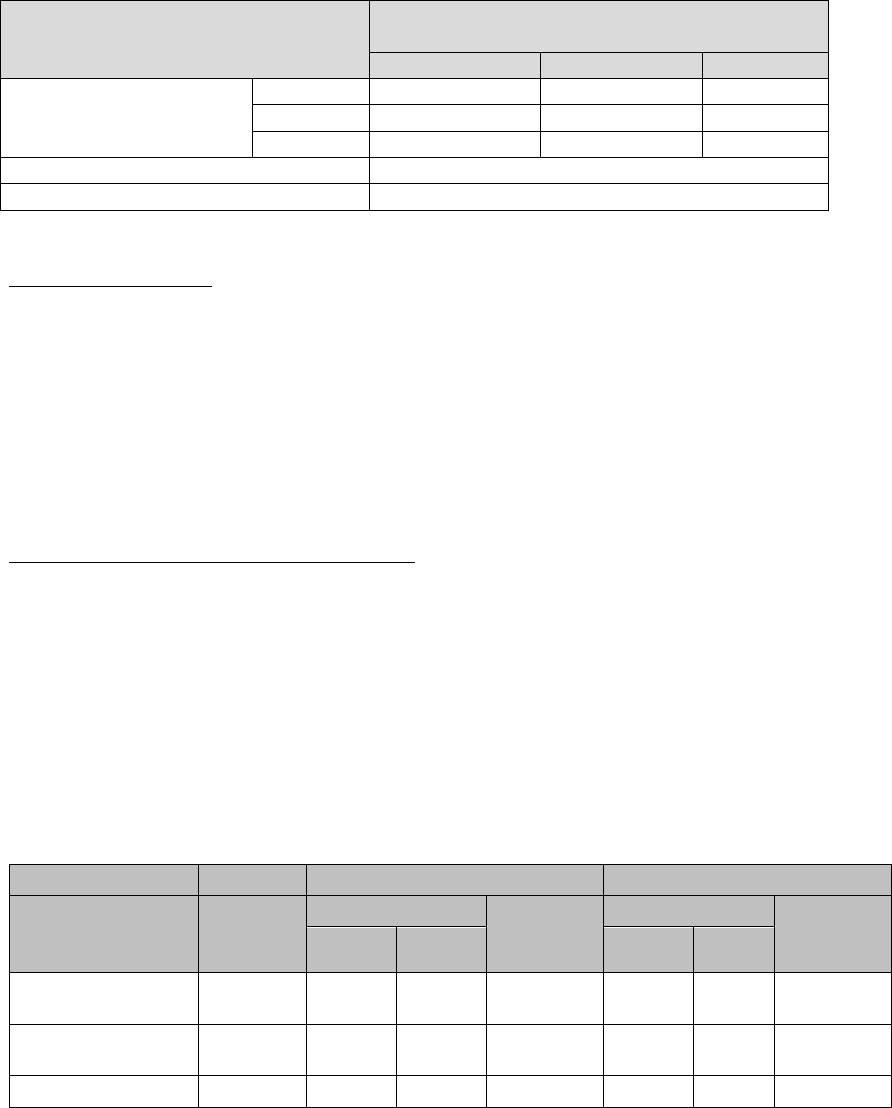
Table 5. Summary of Qualitative Clinical Study Results Performed on the
ABI7500 Fast System
Unmodified CDC EUA Authorized Assay -
Comparator
Positive
Negative
Total
Inform Diagnostics
SARS-CoV-2 RT-PCR
Assay
Positive
31
0
31
Negative
2
31
33
Total
33
31
65
Positive Agreement
93.94% (31/33); 80.40-98.32%
1
Negative Agreement
100.00% (31/31); 88.65-100.00%
1
Two-sided 95% score confidence intervals
Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
9
Table 6. Summary of Qualitative Clinical Study Results Performed on the
QuantStudio Instrument
Unmodified CDC EUA Authorized Assay -
Comparator
Positive
Negative
Total
Inform Diagnostics
SARS-CoV-2 RT-PCR
Assay
Positive
32
0
32
Negative
1
31
32
Total
33
31
64
Positive Agreement
96.97% (32/33); 84.68-99.46%
1
Negative Agreement
100.00% (31/31); 88.65-100.00%
1
Two-sided 95% score confidence intervals
Discordant Analysis:
The discordant samples for the 3 false negative results generated between both
platforms were investigated. It was determined that 2/3 discordant specimens
produced late Ct values (~36 or higher) with the unmodified CDC assay, suggesting
that these specimens may have had target levels below the LoD of the Inform
Diagnostics SASR-CoV-2 RT-PCR Assay. For the third discordant, the investigation
found that an insoluble pellet was present in the extracted RNA from this sample,
which could have had an inhibitory effect on the RT-PCR process and subsequently
led to the false negative result.
Additional Contrived Clinical Evaluation:
30 contrived positive samples were prepared by spiking synthetic viral RNA from
Twist Bioscience into negatively screened nasopharyngeal swab matrix at 1.5X and
2X LoD. The clinical matrix used for spiking was screened negative using the
unmodified CDC assay that Inform Diagnostics uses in-house. In addition, 30
negative clinical matrix samples were also tested. Samples were blinded and
randomized for testing and RNA was extracted using the ViralXpress Reagent.
Testing was performed in one RT-PCR run on both the ABI7500 and QuantStudio
instruments with one positive, one negative, and one extraction control. Results of the
study are summarized below (Table 7).
Table 7. Contrived Clinical Evaluation Summary Data
ABI7500
QuantStudio
SARS-CoV-2
concentration
(copies/µL)
Number
of
samples
Average Ct
Detection
Rate
Average Ct
Detection
Rate
N1 N2 N1 N2
2X LoD
(40 copies/µL)
15 27.83 33.70 15/15 32.43 36.57 15/15
1.5X LoD
(30 copies/µL)
15 30.52 33.94 15/15 33.87 38.81 15/15
Negative
30
UND
UND
0/30
UND
UND
0/30
UND: Undetermined
The results at all tested levels demonstrated 100% agreement and all negative samples
were non-reactive for all SARS-CoV-2 assay targets.

Inform Diagnostics SARS-CoV-2 RT-PCR Assay EUA Summary
10
Clinical Confirmation:
In addition, the first 5 positive and 5 negative samples determined by the Inform
Diagnostics SARS-CoV-2 RT-PCR Assay were sent to the Arizona Health
Department running the CDC EUA test for confirmatory testing. All 10 patient
specimens yielded concordant results.
WARNINGS:
• This test has not been FDA cleared or approved;
• This test has been authorized by FDA under an EUA for use by the authorized
laboratory;
• This test has been authorized only for the detection of nucleic acid from
SARSCoV-2, not for any other viruses or pathogens; and
• This test is only authorized for the duration of the declaration that circumstances
exist justifying the authorization of emergency use of in vitro diagnostics for
detection and/or diagnosis of COVID-19 under Section 564(b)(1) of the Act, 21
U.S.C. § 360bbb-3(b)(1), unless the authorization is terminated or revoked
sooner.
