
For Research Use Only. Not for use in diagnostic procedures.
Novex
™
Pre-Cast gel electrophoresis guide
USER GUIDE
General information and protocols for using Novex
™
pre-cast
gels
Publication Number MAN0003187
Revision A.0
Life Technologies Corporation | 5781 Van Allen Way | Carlsbad, CA 92008
For descriptions of symbols on product labels or product documents, go to thermofisher.com/symbols-definition.
The information in this guide is subject to change without notice.
DISCLAIMER: TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE
LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT, PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR
ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT.
Revision history: Pub. No. MAN0003187
Revision Date Description
A.0 03 March 2020 Updated to current standards.
- 29 October 2010 Baseline for revision.
Important Licensing Information: These products may be covered by one or more Limited Use Label Licenses. By use of these
products, you accept the terms and conditions of all applicable Limited Use Label Licenses.
TRADEMARKS: All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.
©2020 Thermo Fisher Scientific Inc. All rights reserved.
Contents
■
General information ........................................................ 9
Purpose of the guide ............................................................ 9
Storage and shelf life ............................................................ 9
Packaging ...................................................................... 9
Handling the gels .............................................................. 10
Overview of electrophoresis ..................................................... 10
Introduction ............................................................... 10
Support matrix ............................................................ 10
Polyacrylamide gel electrophoresis (PAGE) .................................... 10
Buffer systems ............................................................ 11
Electrophoresis sample conditions ........................................... 11
Power supply considerations for electrophoresis .............................. 11
■
Novex
™
Pre-Cast gels ................................................... 13
Novex
™
gel specications ....................................................... 13
Introduction ............................................................... 13
Specications ............................................................. 13
Novex
™
gel formulations .................................................... 14
Gel selection .................................................................. 16
Choosing a gel for your application .......................................... 16
Protein separation applications .............................................. 16
Nucleic acid separation applications ......................................... 16
Well volume ................................................................... 18
Recommended loading volumes ............................................. 18
Choosing the appropriate well for your application ............................. 18
Gel staining ................................................................... 19
Staining Novex
™
Pre-Cast gels .............................................. 19
■
Methods ................................................................... 20
General guidelines for preparing samples and buffers ............................... 20
Introduction ............................................................... 20
Recommended buffers ..................................................... 20
Reducing agent ........................................................... 21
Running reduced and Non-Reduced samples ................................. 21
Novex
™
Pre-Cast gel electrophoresis guide User Guide
3
Heating samples ........................................................... 21
High salt concentration in samples ........................................... 21
Guanidine-HCl in samples .................................................. 21
Cell lysates ............................................................... 21
Tris-Glycine gels ............................................................... 22
Tris-Glycine discontinuous buffer system ..................................... 22
Materials supplied by the user ............................................... 22
Preparing running buffer .................................................... 23
Preparing samples for denaturing electrophoresis .............................. 23
Preparing samples for native electrophoresis .................................. 23
Electrophoresis conditions .................................................. 24
Staining the gel ............................................................ 24
Tricine gels .................................................................... 25
Tricine buffer system ....................................................... 25
Advantages of tricine gels .................................................. 25
Materials supplied by the user ............................................... 25
Preparing running buffer .................................................... 26
Preparing samples ......................................................... 26
Electrophoresis conditions .................................................. 26
Staining the gel ............................................................ 26
Zymogram gels ................................................................ 27
Zymogram technique ...................................................... 27
Types of zymogram gels .................................................... 27
Materials supplied by the user ............................................... 27
Preparing running buffer .................................................... 28
Preparing samples ......................................................... 28
Electrophoresis conditions .................................................. 28
Detecting protease activity .................................................. 29
Preparing renaturing buffer .................................................. 29
Preparing developing buffer ................................................. 29
Developing zymogram gels ................................................. 29
Staining zymogram gels .................................................... 30
IEF gels ....................................................................... 31
Isoelectric focusing (IEF) .................................................... 31
2D electrophoresis ......................................................... 31
Power considerations for IEF ................................................ 31
Materials supplied by the user ............................................... 32
Preparing anode running buffer (Lower buffer chamber) ........................ 32
Preparing cathode running buffer (Upper buffer chamber) ....................... 32
Preparing sample .......................................................... 33
Add anode and cathode running buffers ...................................... 33
Electrophoresis conditions .................................................. 33
Fixing the gel .............................................................. 33
Staining IEF gels ........................................................... 33
2D SDS-PAGE with IEF gels ................................................. 34
Contents
4
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Materials supplied by the user ............................................... 34
Equilibrating the gel ........................................................ 34
2D separation of proteins on Novex
™
IEF gels ................................. 35
Electrophoresis conditions .................................................. 36
Staining the gel ............................................................ 36
ZOOM
™
gels .................................................................. 37
ZOOM
™
gels .............................................................. 37
2D separation of IPG strips ................................................. 37
Materials supplied by the user ............................................... 37
Equilibrating the IPG strip ................................................... 37
SDS-PAGE ................................................................ 38
Electrophoresis conditions .................................................. 38
Staining the gel ............................................................ 38
TBE gels ...................................................................... 39
Introduction ............................................................... 39
Advantages of TBE gels .................................................... 39
Materials supplied by the user ............................................... 39
Preparing running buffer .................................................... 39
Preparing samples ......................................................... 40
Electrophoresis conditions .................................................. 40
Migration of the dye fronts .................................................. 40
Staining the gel ............................................................ 40
TBE-Urea gels ................................................................. 41
Introduction ............................................................... 41
Materials supplied by the user ............................................... 41
Preparing running buffer .................................................... 41
Preparing samples ......................................................... 42
Electrophoresis conditions .................................................. 42
Migration of the dye fronts .................................................. 42
Staining the gel ............................................................ 42
DNA retardation gels ........................................................... 43
Gel-Shift assay ............................................................ 43
Materials supplied by the user ............................................... 43
Preparing samples ......................................................... 43
Preparing running buffer .................................................... 44
Electrophoresis conditions .................................................. 44
Staining the gel ............................................................ 44
Electrophoresis of Novex
™
Pre-Cast gels .......................................... 45
Introduction ............................................................... 45
Protocol using XCell
™
Mini-Cell
™
............................................ 45
Power supply settings for Novex
™
Pre-Cast gels ................................... 47
Electrophoresis conditions .................................................. 47
Opening Novex
™
Pre-Cast gel cassettes .......................................... 49
Removing the gel after electrophoresis ....................................... 49
Contents
Novex
™
Pre-Cast gel electrophoresis guide User Guide
5
Coomassie
™
staining ........................................................... 50
Introduction ............................................................... 50
Molecular weight calibration ................................................ 50
Materials supplied by the user ............................................... 50
SimplyBlue
™
SafeStain
™
protocol ............................................ 51
Colloidal blue staining kit protocol ........................................... 52
Coomassie
™
R-250 staining protocol ......................................... 52
Silver staining ................................................................. 54
Introduction ............................................................... 54
Molecular weight calibration ................................................ 54
Materials supplied by the user ............................................... 54
Preparing solutions for SilverQuest
™
silver staining ............................. 55
SilverQuest
™
microwave silver staining protocol ............................... 56
Preparing solutions for SilverXpress
™
silver staining ............................ 57
SilverXpress
™
silver staining protocol ......................................... 58
SYPRO
®
Ruby staining ......................................................... 60
Introduction ............................................................... 60
Advantages of SYPRO
®
Ruby staining ........................................ 60
Molecular weight calibration ................................................ 60
Materials supplied by the user ............................................... 60
Preparing solutions for SYPRO
®
Ruby staining ................................ 61
SYPRO
®
Ruby basic protocol ............................................... 61
Visualization of SYPRO
®
Ruby stained gels ................................... 62
Using SYPRO
®
Ruby stain as a Post-Stain .................................... 62
SYBR
™
green staining .......................................................... 63
Introduction ............................................................... 63
Procedure ................................................................ 63
Visualization of SYBR
™
green I stained gels ................................... 63
Ethidium bromide staining ....................................................... 64
Introduction ............................................................... 64
Procedure ................................................................ 64
Gel drying ..................................................................... 65
Introduction ............................................................... 65
Materials supplied by the user ............................................... 65
DryEase
™
Mini-Gel drying system ............................................ 65
Vacuum drying ............................................................ 66
Blotting Novex
™
Pre-Cast gels ................................................... 68
Introduction ............................................................... 68
Power considerations for blotting ............................................ 68
Materials supplied by the user ............................................... 68
Preparing transfer buffer .................................................... 69
Preparing transfer buffer for TBE gels ........................................ 69
Preparing transfer buffer compatible with protein sequencing ................... 69
Preparing blotting pads ..................................................... 69
Preparing transfer membrane and lter paper ................................. 70
Contents
6
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Western transfer using the XCell II blot module ................................ 70
Recommended transfer conditions ........................................... 73
Blotting IEF gels ........................................................... 74
Blotting native gels ........................................................ 74
Calibrating protein molecular weight .............................................. 75
Introduction ............................................................... 75
Protein secondary structure ................................................. 75
Buffer systems ............................................................ 75
Assigned apparent molecular weights ........................................ 76
Troubleshooting ................................................................ 78
■
APPENDIX A Appendix ................................................ 80
Accessory products ............................................................ 80
Electrophoresis reagents ................................................... 80
Protein stains and standards ................................................ 82
Nucleic acid markers ....................................................... 83
Recipes ....................................................................... 84
Tris-Glycine SDS running buffer ............................................. 84
Tris-Glycine native running buffer ............................................ 84
Tris-Glycine SDS sample buffer .............................................. 85
Tris-Glycine native sample buffer ............................................ 85
Tris-Glycine transfer buffer .................................................. 86
Tricine SDS sample buffer .................................................. 86
Tricine SDS running buffer .................................................. 87
10X zymogram renaturing buffer ............................................. 87
Zymogram developing buffer ................................................ 87
IEF sample buffer pH 3–7 ................................................... 88
IEF sample buffer, pH 3–10 ................................................. 88
IEF cathode buffer, pH 3–7 .................................................. 89
IEF cathode buffer, pH 3–10 ................................................. 89
IEF anode buffer ........................................................... 89
TBE running buffer ......................................................... 89
Hi-Density TBE sample buffer ............................................... 90
TBE-Urea sample buffer .................................................... 90
Prep TBE–Urea sample buffer ............................................... 91
Gel migration charts ............................................................ 92
Novex
™
Tris-Glycine gel migration chart ...................................... 92
Novex
™
tricine, IEF, and zymogram gel migration chart ......................... 93
Novex
™
TBE and TBE-Urea gel migration chart ................................ 94
ZOOM
™
gel migration chart ................................................. 95
References .................................................................... 96
Contents
Novex
™
Pre-Cast gel electrophoresis guide User Guide
7
■
APPENDIX B Safety .................................................... 97
Chemical safety ................................................................ 98
Biological hazard safety ......................................................... 99
■
Documentation and support ........................................... 100
Customer and technical support ................................................ 100
Limited product warranty ...................................................... 100
Contents
8
Novex
™
Pre-Cast gel electrophoresis guide User Guide

General information
WARNING! Read the Safety Data Sheets (SDSs) and follow the handling
instructions. Wear appropriate protective eyewear, clothing, and gloves. Safety Data
Sheets (SDSs) are available from thermofisher.com/support.
Purpose of the guide
We have available a variety of pre-cast gels for use with the XCell
™
Mini-Cell
™
. These
include gels for analysis of proteins (Tris-Glycine, Tricine, Zymogram, IEF, and
ZOOM
™
Gels) and nucleic acids (TBE, TBE-Urea, and DNA Retardation).
The Novex
™
Pre-Cast Gel Electrophoresis Guide contains information about the
Novex
™
Pre-Cast gels and is intended to supplement the Gel Instruction Cards
(IM-6000 to IM-6008) supplied with the pre-cast gels. Complete protocols for sample
and buffer preparation, electrophoresis conditions, staining, and blotting are provided
in this guide.
Storage and shelf life
Store Novex
™
Pre-Cast Gels at +4℃. The gels have a shelf life of 4–8 weeks
depending upon the gel type when stored at +4℃.
Do not freeze Novex
™
Pre-Cast Gels.
Use gels immediately from the refrigerator. Extended exposure of the gels to room
temperature signicantly impairs the performance of the gel.
Packaging
The Novex
™
Pre-Cast Gels are supplied as 10 gels per box. Gels are individually
packaged in clear pouches with 4–10 mL of Packaging Buffer.
Novex
™
Pre-Cast gel electrophoresis guide User Guide
9

Handling the gels
The Packaging Buffer contains 0.02% sodium azide and residual acylamide
monomer. Wear gloves at all times when handling gels.
WARNING! This product contains a chemical (acrylamide) known to the state of
California to cause cancer.
Overview of electrophoresis
Electrophoresis is a simple, rapid, and sensitive analytical tool for separating proteins
and nucleic acids based on their physical characteristics (mass, isoelectric point,
etc.).
Most biological molecules carry a net charge at any pH other than their isoelectric
point and migrate at a rate proportional to their charge density in an electrical eld.
The mobility of a biological molecule through an electric eld depends on the
following factors:
•
Field strength
•
Net charge on the molecule
•
Size and shape of the molecule
•
Ionic strength
•
Properties of the medium through which the molecules migrate (e.g., viscosity,
pore size)
Polyacrylamide and agarose are two types of support matrices used in
electrophoresis. The support matrix is a porous media that acts as a molecular sieve.
The sieving function depends on the pore size, and concentration of the matrix.
Agarose has a large pore size and is ideal for separating macro-molecules such as
nucleic acids and protein complexes. Polyacrylamide has a smaller pore size and is
ideal for separating proteins and smaller nucleic acids.
Polyacrylamide gels are formed by the polymerization of acrylamide monomers into
long chains, crosslinked by bifunctional compounds such as N,N-methylene-
bisacrylamide (bis) that react with the free functional groups at the chain termini.
The pore size of the gel is governed by the concentration of acrylamide and
bisacrylamide (%T and %C).
%T = concentration of total monomer
%C = proportion of cross linker (as a percentage of total monomer)
The higher the acrylamide concentration, the smaller the pore size, allowing
resolution of low molecular weight molecules and vice-versa.
Introduction
Support matrix
Polyacrylamide
gel
electrophoresis
(PAGE)
General information
Handling the gels
10
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Electrophoresis is performed using continuous or discontinuous buffer systems.
Continuous buffer systems utilize a single buffer for the gel and the running buffer.
Discontinuous buffer systems (Ornstein 1964) utilize different gel buffers and running
buffer. In addition, two gel layers of different pore size, the stacking and separating
gel, are used. Electrophoresis using a discontinuous buffer system allows
concentration of the sample to a narrow region prior to separation, resulting in
sharper bands and higher resolution.
Depending upon the application, electrophoresis can be performed under the
following conditions:
Denaturing
Electrophoresis is performed under denaturing conditions using an anionic detergent
such as sodium dodecylsulfate (SDS). SDS denatures and unfolds the proteins by
binding the hydrophobic portions of the protein at a ratio of ∼1.4 g SDS per gram of
protein. The resultant SDS-protein complexes are highly negatively charged and
migrate through the gel based on their size rather than charge.
Non-Denaturing (Native)
Electrophoresis is performed under non-denaturing (native) conditions using buffer
systems that maintain the native protein conformation, cohesion of subunits, and
biological activity. During native electrophoresis, proteins are separated based on
their charge to mass ratios.
Reducing
Electrophoresis is performed under reducing conditions using reducing agents such
as dithiothreitol (DTT) or β-mercaptoethanol (β-ME). The reducing agents cleave any
disulde bonds between cysteine residues resulting in complete separation of
denatured proteins into their individual subunits.
In electrical terms, the process of electrophoresis is closely associated with the
following equations derived from Ohm′s Law:
Voltage = Current × Resistance (V=IR)
Wattage = Current × Voltage (W=IV)
Resistance
The electrical resistance of the assembled electrophoresis cell is dependent on buffer
conductivity, gel thickness, temperature, and the number of gels being run. Although
the resistance is determined by the gel system, the resistance varies over the course
of the run.
•
In discontinuous buffer systems (and to a lesser extent in continuous buffer
systems) resistance increases over the course of electrophoresis. This occurs in
the Tris-Glycine buffer system as highly conductive chloride ions in the gel are
replaced by less conductive glycine ions from the running buffer.
•
Resistance decreases as the temperature increases.
Voltage
Buffer systems
Electrophoresis
sample
conditions
Power supply
considerations
for
electrophoresis
General information
Overview of electrophoresis
Novex
™
Pre-Cast gel electrophoresis guide User Guide
11

The velocity of an ion in an electric eld varies in proportion to the eld strength (Volts
per unit distance). The higher the voltage, the faster an ion moves. For most
applications, we recommend a constant voltage setting.
•
A constant voltage setting allows the current and power to decrease over the
course of electrophoresis, providing a safety margin in case of a break in the
system.
•
The constant voltage setting does not need adjustment to account for
differences in number or thickness of gels being electrophoresed.
Current
For a given gel/buffer system, at a given temperature, current varies in proportion to
the eld strength (voltage) and/or cross-sectional area (thickness and/or number of
gels). When using a constant current setting, migration starts slow, and accelerates
over time, thus favoring stacking in discontinuous gels.
When running under constant current, set a voltage limit on the power supply at, or
slightly above the maximum expected voltage to avoid unsafe conditions. At
constant current voltage increases as resistance increases. If a local fault condition
occurs (e.g., a bad connection), high local resistance may cause the voltage to reach
the maximum for the power supply, leading to overheating and damage of the
electrophoresis cell.
Power
Wattage measures the rate of energy conversion, which is manifest as heat
generated by the system. Using constant power ensures that the total amount of
heat generated by the system remains constant throughout the run, but results in
variable mobility since voltage increases and current decreases over the course of
the run. Constant power is typically used when using IEF strips.
When using constant power, set the voltage limit slightly above the maximum
expected for the run. High local resistance can cause a large amount of heat to be
generated over a small distance, damaging the electrophoresis cell and gels.
General information
Overview of electrophoresis
12
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Novex
™
Pre-Cast gels
Novex
™
gel specifications
The Novex
™
Pre-Cast Gel cassette is 10 cm × 10 cm in size, and designed for use
with the XCell
™
Mini-Cell
™
and XCell6
™
MultiGel Unit (see “Accessory products” on
page 80 for ordering information).
Novex
™
Pre-Cast Gels are available for resolving proteins in the range of 2–500 kDa
and nucleic acids in the range of 10–3,000 bp, depending upon the type and
acrylamide percentage of the gel. Refer to Gel Selection (“Choosing a gel for your
application” on page 16) for details on applications and migration patterns.
Gel Matrix
™
:
Acrylamide/Bisacrylamide
Gel Thickness: 1.0 mm or 1.5 mm
Gel Size: 8 cm × 8 cm
Cassette Size: 10 cm × 10 cm
Cassette Material: Styrene Copolymer (recycle code 7)
Sample Well Conguration: 1, 5, 9, 10, 12, 15-well, 2D-well, and IPG
well
Introduction
Specifications
Novex
™
Pre-Cast gel electrophoresis guide User Guide
13
All Novex
™
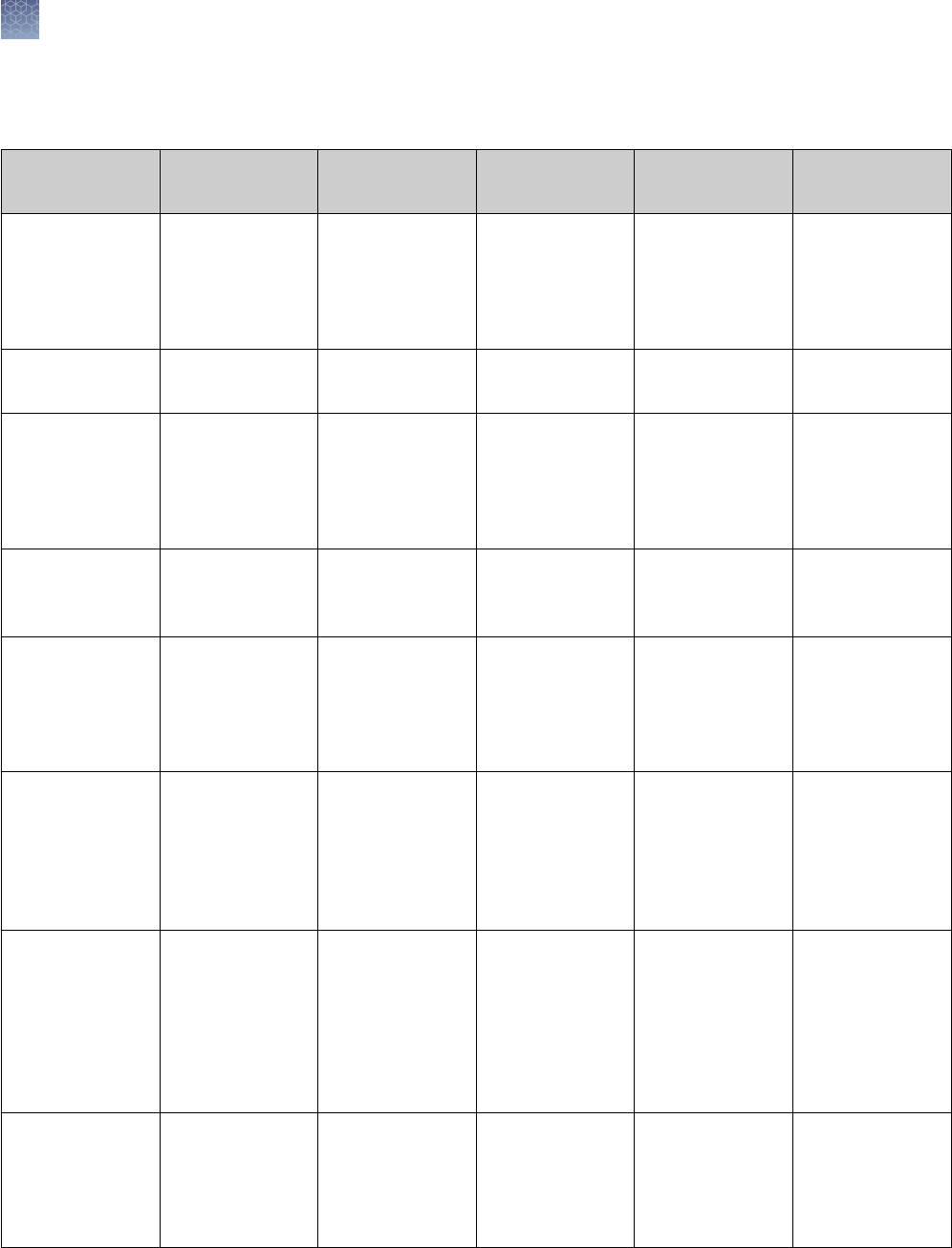
Pre-Cast gels are made with high purity reagents. The gels for DNA
analysis are DNase-free. The composition of the different gels is listed below:
Gel Type Formulation Stacking Gel Separating Gel
% Bis-
Acrylamide
pH
Tris-Glycine Gels
(except 4%)
Tris-base, HCl,
Acrylamide, Bis-
acrylamide,
TEMED, APS,
Ultrapure water
4% 6%, 8%, 10%,
12%, 14%, 16%,
18%, 4–12%, 8–
16%, 4–20%, 10–
20%
2.6% 8.6
4% Tris-Glycine
Gels
Same as Tris
Glycine
3.5% 4% 1.3% 8.6
Tricine Gels Tris-base, HCl,
Acrylamide, Bis-
acrylamide,
TEMED, APS,
Ultrapure water
4% 10%, 16%, 10–
20%
2.6% 8.3
Zymogram Gels Tris Glycine Gels
with a substrate,
casein or gelatin
4%
No substrate
10%, 12%, 4–
16%
2.6% 8.6
IEF Gels Acrylamide, Bis-
acrylamide,
TEMED, APS,
Ultrapure water,
2% ampholytes
None pH 3–7 pH 3–10 2.6% 5.0 6.0
TBE Gels Tris-base, Boric
acid, EDTA,
Acrylamide, Bis-
acrylamide,
TEMED, APS,
Ultrapure water
4% 6%, 8%, 10%,
20%, 4–12%, 4–
20%
2.6% 8.3
TBE-Urea Gels Tris-base, Boric
acid, EDTA,
Acrylamide, Bis-
acrylamide,
TEMED, APS,
Ultrapure water,
7M Urea
4% 6%, 10%, 15% 3.8–5% 8.7
DNA Retardation
Gels
6%
polyacrylamide
gels prepared
with half strength
TBE gel buffer
None 6% 2.6% 8.3
Novex
™
gel
formulations
Novex
™
Pre-Cast gels
Novex
™
gel specifications
14
Novex
™
Pre-Cast gel electrophoresis guide User Guide

IMPORTANT! Novex
™
Pre-Cast gels do not contain SDS. These gels can be used
for non-denaturing (native) and denaturing gel electrophoresis.
For optimal and total separation ranges for each specic gel percentage, consult the
Gel Migration Charts on (“Gel migration charts” on page 92).
Novex
™
Pre-Cast gels
Novex
™
gel specifications
Novex
™
Pre-Cast gel electrophoresis guide User Guide
15

Gel selection
To obtain the best results, it is important to choose the correct gel percentage, buffer
system, gel format, and thickness for your application.
Review the following section, and Well Volume (“Well volume” on page 18) to
determine the type of gel that is best suited for your application.
Refer to the Novex
™
Gel Migration Charts (see “Gel migration charts” on page 92) to
nd the gel with the region of maximum resolution best suited for your sample. The
leading protein molecules should migrate about 70% of the length of gel for best
resolution.
Separation of proteins over a wide range of molecular weights
Use Novex
™
Tris-Glycine Gels for separating proteins over a wide molecular weight
range (6–200 kDa) under denaturing or non-denaturing conditions.
Resolve large molecules with low percentage gels, and small molecules with high
percentage gels. If the molecular weight of the molecule is unknown, or the sample
contains a wide range of molecules, use a gradient gel.
Separation of low molecular weight proteins and peptides
The Novex
™
Tricine Gels provide high resolution of low molecular weight proteins and
peptides (2–200 kDa). Tricine gels give the best results under denaturing conditions.
Isoelectric focusing (IEF)
Use Novex
™
IEF Gels for native (vertical) IEF of proteins. The pH 3–10 gels have a pI
performance range of 3.5–8.5 and pH 3–7 gels have a pI performance range of 3.0–
7.0.
Protease detection
The Novex
™
Zymogram Gels are used for detecting and characterizing proteases that
utilize casein or gelatin as the substrate. Proteins are run under denaturing conditions
and then renatured for enzymatic activity.
2D separation of proteins
The ZOOM
™
Gels are specically designed for second dimension electrophoresis of
7.0 cm IPG strips. Gels with 2D wells can also be used, but only accommodate IPG
strips of 6.5 cm.
Nucleic acid analysis
The Novex
™
Pre-Cast Gels are capable of resolving nucleic acids in the range of 10–
3000 bp.
Novex
™
TBE Gels are used to perform analysis of DNA fragments from restriction
digest and PCR products, Southern analysis, and primer analysis.
Novex
™
TBE-Urea Gels are used for denaturing nucleic acid analysis and are suited
for RNase Protection Assays, in-vitro transcription studies, RNA stability studies, and
oligonucleotide purication.
Choosing a gel
for your
application
Protein
separation
applications
Nucleic acid
separation
applications
Novex
™
Pre-Cast gels
Gel selection
16
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Gel shift assays
The Novex
™
6% DNA Retardation Gels are used to perform gel shift assays.
Novex
™
Pre-Cast gels
Gel selection
Novex
™
Pre-Cast gel electrophoresis guide User Guide
17
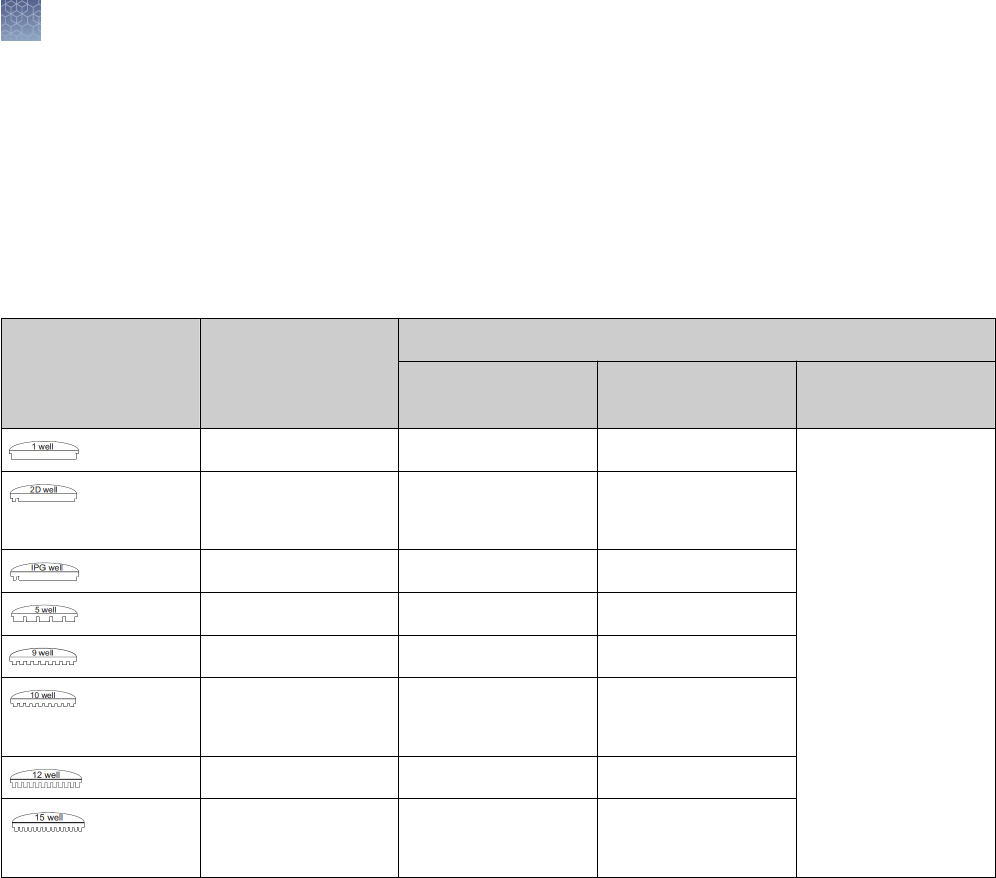
Well volume
The recommended loading volumes and protein load per band by the detection
method are provided in the table below.
Note: The 9-well gels are compatible with any eight-channel pipettors used for
loading samples from 96-well plates. An additional lane is included for loading
protein molecular weight standard.
Well Types
Maximum Load
Volume
Maximum Protein Load Per Band by Detection Method
Coomassie
™
Staining
Ethidium Bromide Silver Staining
1.0 mm 700 µL 12 µg/band 2.4 µg/band Scale your sample
load for the sensitivity
of your silver staining
kit.
For use with the
SilverQuest
™
or
SilverXpress
™
Silver
Staining Kits, we
recommend a protein
load of 1 ng/band.
1.0 mm
1.5 mm
400 µL
600 µL
12 µg/band 2.0 µg/band
1.0 mm 7 cm IPG Strip N/A N/A
1.0 mm 60 µL 2 µg 400 ng/band
1.0 mm 28 µL 0.5 µg/band 100 ng/band
1.0 mm
1.5 mm
25 µL
37 µL
0.5 µg/band 100 ng/band
1.0 mm 20 µL 0.5 µg/band 100 ng/band
1.0 mm
1.5 mm
15 µL
25 µL
0.5 µg/band 100 ng/band
Choose the type of well for your application based upon the volume of your sample.
The more wells a comb has, and the thinner the gel is, the lower the sample loading
volume.
Note: Proteins transfer out of a 1.0 mm gel more easily than from a 1.5 mm gel.
Recommended
loading
volumes
Choosing the
appropriate
well for your
application
Novex
™
Pre-Cast gels
Well volume
18
Novex
™
Pre-Cast gel electrophoresis guide User Guide
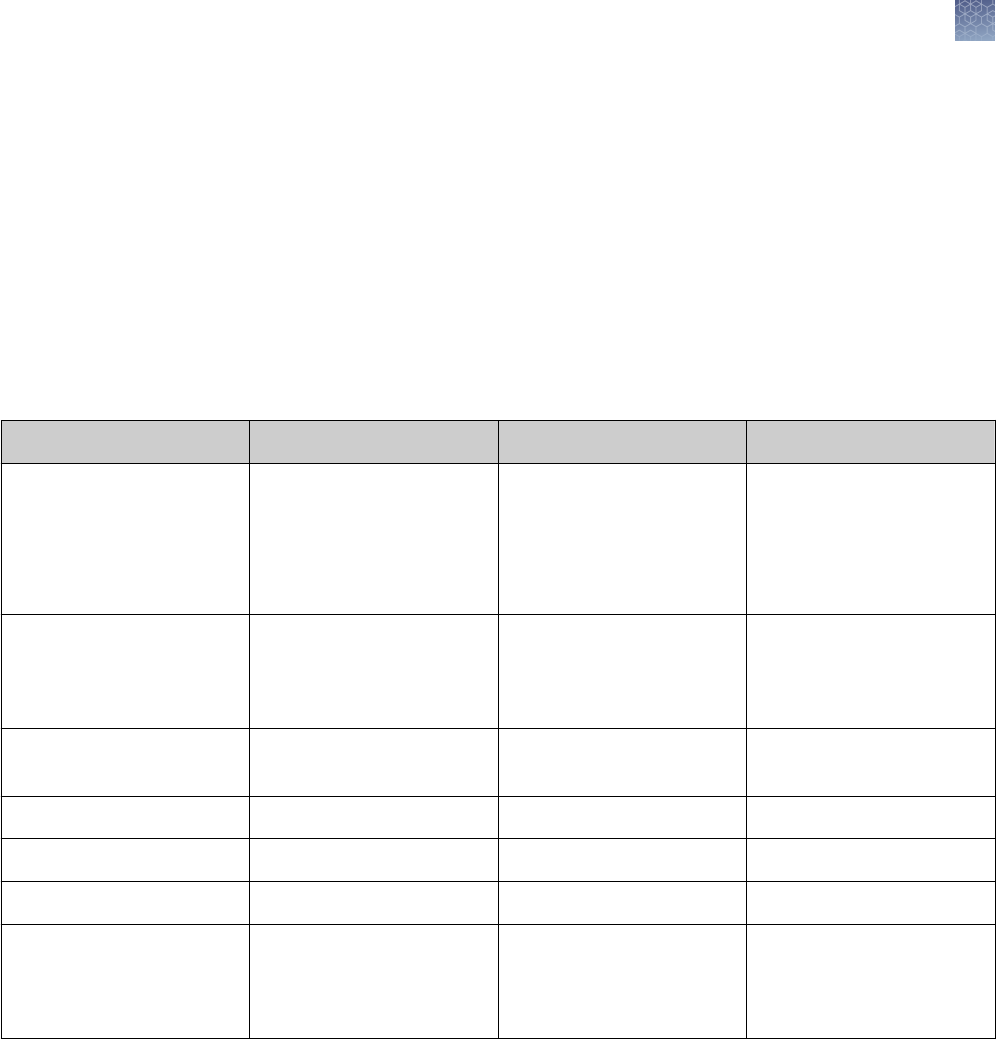
Gel staining
The Novex
™
Pre-Cast Gels are compatible with most silver staining protocols. We
recommend using the SilverQuest
™
Silver Staining Kit or the SilverXpress
™
Silver
Staining Kit (see “Silver staining” on page 54) for silver staining of Novex
™
Gels.
Novex
™
Pre-Cast Gels are compatible with any of the standard Coomassie
™
staining
procedures. Protocols that are accelerated by heat are preferable, as heat can x
proteins (especially smaller peptides). The SimplyBlue
™
SafeStain
™
(see
“SimplyBlue
™
SafeStain
™
protocol” on page 51) and Novex
™
Colloidal Blue Staining
Kit (see “Colloidal blue staining kit protocol” on page 52) are recommended for
staining Novex
™
Gels.
Stain Type Sensitivity Gel Type Compatibility Application
Coomassie
™
Blue
Coomassie
™
Fluor
™
Orange
Colloidal Coomassie
™
Blue
SimplyBlue
™
SafeStain
™
100–500 ng
8–16 ng
<10 ng
5 ng
Tris-Glycine, Bis-Tris,
Tricine, native
General
SilverXpress
™
SilverQuest
™
1 ng
0.3–2.5 ng
0.3–0.9 ng (50 bp)
Tris-Glycine, Bis-Tris,
Tricine, TBE
Bis-Tris, Tricine, TBE
Low sample quantity,
Nucleic acid
SYPRO
®
Ruby 0.25–1 ng Tris-Glycine, Bis-Tris,
Tricine, native
Low sample quantity,
Nucleic acid, Mass Spec
Pro-Q
™
Diamond 1–16 ng Tris-Glycine, Bis-Tris Phosphoprotein
Pro-Q
™
Emerald 0.5–3 ng Tris-Glycine Glycoprotein
Ethidium Bromide 10 ng (50 bp) TBE Nucleic acid
SYBR
™
Green 60 pg (dsDNA)
100–300 pg (ssDNA)
1–2 ng (24 bp)
TBE Nucleic acid
Staining
Novex
™
Pre-
Cast gels
Novex
™
Pre-Cast gels
Gel staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
19
Methods
General guidelines for preparing samples and buffers
The XCell
™
Mini-Cell
™
and a power supply are needed to perform electrophoresis
with Novex
™
Pre-Cast gels. Additional reagents supplied by the user are described
for each individual protocol.
General guidelines for preparing samples and buffers for Novex
™
Pre-Cast gels are
discussed below. Detailed instructions for preparing the sample buffer and running
buffer are described in the sections for each individual type of gel.
The recommended running buffer and sample buffer for each Novex
™
Pre-Cast Gel is
listed in the table below. Prepare your sample in the appropriate sample buffer such
that the nal concentration of the sample buffer is 1X.
Running buffer must be diluted to 1X nal concentration before use.
See “Accessory products” on page 80 for ordering information on pre-mixed
buffers. See “Tris-Glycine SDS running buffer” on page 84 for recipes if you are
preparing your own buffers.
Gel Type
Running Buffer Sample Buffer
Novex
™
Tris-Glycine Gels (SDS-
PAGE)
Tris-Glycine SDS Running Buffer
(10X)
Tris-Glycine SDS Sample Buffer (2X)
Novex
™
Tris-Glycine Gels (Native-
PAGE)
Tris-Glycine Native Running Buffer
(10X)
Tris-Glycine Native Sample Buffer
(2X)
Novex
™
Tricine Gels Tricine SDS Running Buffer (10X) Tricine SDS Sample Buffer (2X)
Novex
™
Zymogram Gels Tris-Glycine SDS Running Buffer
(10X)
Tris-Glycine SDS Sample Buffer (2X)
IEF Gels IEF Cathode Buffer (10X)
IEF Anode Buffer (50X)
IEF Sample Buffer (2X)
TBE Gels TBE Running Buffer (5X) Hi-Density TBE Sample Buffer (5X)
TBE-Urea Gels TBE Running Buffer (5X) TBE-Urea Sample Buffer (2X)
Prep TBE-Urea Sample Buffer (2X)
for preparative gels
DNA Retardation Gels TBE Running Buffer (5X) Hi-Density TBE Sample Buffer (5X)
Introduction
Recommended
buffers
20
Novex
™
Pre-Cast gel electrophoresis guide User Guide

When preparing samples for reducing gel electrophoresis, any of the following
reducing agents may be used:
•
NuPAGE
™
Reducing Agent (see “Accessory products” on page 80 for ordering
information)
•
Dithiothreitol (DTT), 50 mM nal concentration
•
β-mercaptoethanol, 2.5% nal concentration
•
tris(2-carboxyethyl)phosphine (TCEP), 50 mM nal concentration
Add the reducing agent to the sample up to an hour before loading the gel.
Avoid storing reduced samples for long periods, even if they are frozen. Reoxidation
of samples occur during storage and produce inconsistent results.
For optimal results, we do not recommend running reduced and non-reduced
samples on the same gel.
If you do choose to run reduced and non-reduced samples on the same gel, do not
run reduced and non-reduced samples in adjacent lanes. The reducing agent may
have a carry-over effect on the non-reduced samples if they are in close proximity.
Heating the sample at 100℃ in SDS containing buffer results in proteolysis (Kubo,
1995). We recommend heating samples for denaturing electrophoresis (reduced or
non-reduced) at 85℃ for 2–5 minutes for optimal results.
Do not heat the samples for non-denaturing (native) electrophoresis or
Zymogram Gels.
High salt concentrations result in increased conductivity that affects protein
migration, and can result in gel artifacts in adjacent lanes containing samples with
normal salt concentrations. Perform dialysis or precipitate and resuspend samples in
lower salt buffer prior to electrophoresis.
Samples solubilized in guanidine-HCl have high ionic strength, and produce
increased conductivity similar to high salt concentrations. In addition, guanidine
precipitates in the presence of SDS leading to various types of gel artifacts. If
possible, change the solubilization agent by dialysis prior to electrophoresis.
Take the following considerations into account when performing electrophoresis of
cell lysates:
•
Genomic DNA in the cell lysate may cause the sample to become viscous and
affect protein migration patterns and resolution. Shear genomic DNA to reduce
viscosity before loading the sample.
•
Cells lysates contain soluble and insoluble fractions. The size of each fraction
depends upon the type of sample being analyzed. The nature of the insoluble
fraction may result in altered protein migration patterns and resolution. Separate
the two fractions by centrifugation and load them on separate lanes for
electrophoresis.
•
If RIPA buffer is used in cell lysis, subsequent blotting of proteins <40 kDa may
be inhibited due to the presence of Triton
™
X-100 in the buffer.
Reducing
agent
Running
reduced and
Non-Reduced
samples
Heating
samples
High salt
concentration
in samples
Guanidine-HCl
in samples
Cell lysates
Methods
General guidelines for preparing samples and buffers
Novex
™
Pre-Cast gel electrophoresis guide User Guide
21

Tris-Glycine gels
Novex
™
Tris-Glycine gels are based on the Laemmli System (Laemmli, 1970) with
minor modications for maximum performance in the pre-cast format. Unlike
traditional Laemmli gels with a stacking gel pH of 6.8 and separating gel pH of 8.8,
Novex
™
Tris-Glycine gels have a pH of 8.65 for both regions.
The Tris-Glycine discontinuous buffer systems utilizes three ions:
•
Chloride (
–
) from the gel buffer serves as a leading ion due to its high afnity to
the anode relative to other anions in the system. The gel buffer ions are Tris
+
and
Cl
–
(pH 8.65).
•
Glycine (
–
) is the primary anion in the running buffer and serves as a trailing ion.
Glycine is partially negatively charged and trails behind the highly charged
chloride ions in the charged environment. The running buffer ions are Tris
+
, Gly
–
,
and dodecylsulfate
–
(pH 8.3).
•
Tris Base (
+
) is the common ion present in the gel buffer and running buffer.
During electrophoresis, the gel and buffer ions in the Tris-Glycine system form an
operating pH of 9.5 in the separation region of the gel.
The following reagents are needed to perform electrophoresis with Novex
™
Tris-
Glycine Gels. Ordering information for pre-mixed buffers is on “Accessory products”
on page 80. If you are preparing your own buffers, recipes are provided on
“Recipes” on page 84.
•
Protein sample
•
Deionized water
•
Protein molecular weight markers
For denaturing electrophoresis
•
Novex
™
Tris-Glycine SDS Sample Buffer
•
NuPAGE
™
Reducing Agent
•
Novex
™
Tris-Glycine SDS Running Buffer
For non-denaturing (native) electrophoresis
•
Novex
™
Tris-Glycine Native Sample Buffer
•
Novex
™
Tris-Glycine Native Running Buffer
Tris-Glycine
discontinuous
buffer system
Materials
supplied by the
user
Methods
Tris-Glycine gels
22
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Use 1X Tris-Glycine SDS Running Buffer for electrophoresis of denatured samples, or
1X Native Running Buffer for electrophoresis of native samples.
1.
Prepare 1,000 mL of Running Buffer as described below:
Reagent Amount
10X Novex
™
Tris-Glycine SDS or 10X
Native Running Buffer
100 mL
Deionized Water 900 mL
Total Volume 1,000 mL
2.
Mix the buffer thoroughly and use it to ll the Upper and Lower Buffer Chambers
of the assembled XCell
™
Mini-Cell
™
for electrophoresis.
To separate proteins by mass alone, denature samples using SDS Sample Buffer and
heating.
1.
Prepare each sample as described below:
Reagent Amount
Sample x µL
Novex
™
Tris-Glycine SDS Sample Buffer
(2X)
5 µL
Deionized Water to 5 µL
Total Volume 10 µL
2.
Heat the sample at 85℃ for 2 minutes. Load the samples onto the gel
immediately.
Note: For reduced samples, add the reducing agent to a nal concentration of
1X immediately prior to electrophoresis to obtain the best results.
To separate proteins by charge:mass ratio in their native conformation, use non-
denaturing (native) electrophoresis.
1.
Prepare each sample as described below:
Reagent Amount
Sample x µL
Novex
™
Tris-Glycine Native Sample
Buffer (2X)
5 µL
Deionized Water to 5 µL
Total Volume 10 µL
2.
Load the samples onto the gel immediately. Do not heat samples for native
electrophoresis.
Preparing
running buffer
Preparing
samples for
denaturing
electrophoresis
Preparing
samples for
native
electrophoresis
Methods
Tris-Glycine gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
23

See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
. Run the gel at 125 V
constant. See “Electrophoresis conditions” on page 47 for additional details on
electrophoresis conditions.
Any of the techniques described on “Coomassie
™
staining” on page 50–“Using
SYPRO
®
Ruby stain as a Post-Stain” on page 62 are suitable for staining Novex
™
Tris-Glycine Gels after electrophoresis.
Electrophoresis
conditions
Staining the gel
Methods
Tris-Glycine gels
24
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Tricine gels
The Tricine system is a modication of the Tris-Glycine discontinuous buffer system
(see “Tris-Glycine gels” on page 22) developed by Schaegger and von Jagow
(Schaegger and von Jagow, 1987) specically designed for resolving peptides and
low molecular weight proteins.
In the Tris-Glycine system, proteins are stacked in the stacking gel between the
highly mobile leading chloride ion (in the gel buffer) and the slower trailing glycine ion
(in the running buffer). These stacked protein bands undergo sieving once they reach
the separating gel.
However, the resolution of smaller proteins (<10 kDa) is hindered by the continuous
accumulation of free dodecylsulfate (DS) ions (from the SDS sample and running
buffers) in the stacking gel. Smaller proteins mix with DS ions in the zone of stacked
DS micelles, resulting in fuzzy bands and decreased resolution. The mixing also
interferes with the xing and staining of smaller proteins.
To avoid this problem, the Tricine system uses a low pH gel buffer and replaces the
trailing glycine ion with a fast moving tricine ion in the running buffer. The smaller
proteins that previously migrated with the stacked DS micelles in the Tris-Glycine
system become well separated from DS ions in the Tricine system, resulting in more
efcient stacking and destacking of low molecular weight proteins, sharper bands,
and higher resolution
The Tricine Gels have the following advantages over the Tris-Glycine Gels for
resolving proteins in the molecular weight range of 2–20 kDa:
•
Allows resolution of proteins with molecular weights as low as 2 kDa
•
Ideal for direct sequencing of proteins after transferring to PVDF as tricine does
not interfere with sequencing
•
Minimizes protein modication because of a lower pH
The following reagents are needed to perform electrophoresis with Novex
™
Tricine
Gels. Ordering information for pre-mixed buffers is on “Accessory products” on
page 80. If you are preparing your own buffers, recipes are provided on “Recipes”
on page 84.
•
Protein sample
•
Deionized water
•
Protein molecular weight markers
•
Novex
™
Tricine SDS Sample Buffer
•
NuPAGE
™
Reducing Agent for reduced samples
•
Novex
™
Tricine SDS Running Buffer
Tricine buffer
system
Advantages of
tricine gels
Materials
supplied by the
user
Methods
Tricine gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
25

Use 1X Novex
™
Tricine SDS Running Buffer for electrophoresis of Tricine gels.
1.
Prepare 1,000 mL of Running Buffer as described below:
Reagent Amount
Novex
™
Tricine SDS Running Buffer
(10X)
100 mL
Deionized Water 900 mL
Total Volume 1,000 mL
2.
Mix thoroughly. Use this buffer to ll the Upper and Lower Buffer Chambers of
the XCell
™
Mini-Cell
™
for electrophoresis.
Note: Novex
™
Tricine Gel are not compatible with buffers for Tris-Glycine gels.
·
Samples run in Tris-Glycine SDS Sample Buffer are poorly resolved.
·
Samples run in Tris-Glycine SDS Running Buffer take longer to complete and result
in poor resolution of smaller proteins.
Protein samples for Tricine Gels can be denatured, or denatured and reduced.
1.
Prepare each reduced or non-reduced samples for running on Tricine gels as
described below:
Reagent Reduced Sample Non-reduced Sample
Sample x µL x µL
Novex
™
Tricine SDS
Sample Buffer (2X)
5 µL 5 µL
NuPAGE
™
Reducing Agent
(10X)
1 µL –
Deionized Water to 4 µL to 5 µL
Total Volume 10 µL 10 µL
2.
Heat samples at 85℃ for 2 minutes. Load the samples onto the gel immediately.
Note: For reduced sample, add the reducing agent immediately prior to
electrophoresis to obtain the best results. Leave an empty lane between
samples with and without reducing agent to prevent diffusion of the reducing
agent into non-reduced sample lanes.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
. Run the gel at 125 V
constant. See “Electrophoresis conditions” on page 47 for additional details on
electrophoresis conditions.
Any of the techniques described on “Coomassie
™
staining” on page 50–“Using
SYPRO
®
Ruby stain as a Post-Stain” on page 62 are suitable for staining Novex
™
Tricine Gels after electrophoresis.
Preparing
running buffer
Preparing
samples
Electrophoresis
conditions
Staining the gel
Methods
Tricine gels
26
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Zymogram gels
Zymogram analysis is used for detecting and characterizing metalloproteinases,
collagenases, and other proteases that can utilize casein or gelatin as a substrate.
Protease samples are denatured in SDS buffer under non-reducing conditions and
without heating, and run on a Zymogram Gel using Tris-Glycine SDS Running Buffer.
After electrophoresis, the enzyme is renatured by incubating the gel in Zymogram
Renaturing Buffer containing a non-ionic detergent. The gels are then equilibrated in
Zymogram Developing Buffer (to add divalent metal cations required for enzymatic
activity), and then stained and destained. Regions of protease activity appear as
clear bands against a dark blue background where the protease has digested the
substrate.
Three different types of Zymogram Gels are available from Thermo Fisher Scientic.
Details are listed on the table below.
Gel Type Separating Gel Substrate Sensitivity
Novex
™
Zymogram
Gelatin Gel
10% Tris-Glycine gel with 0.1% gelatin 10
–6
units of
collagenase
Novex
™
Zymogram
Casein Gel
12% Tris-Glycine gel β-casein 7 × 10
–4
units of
trypsin
Novex
™
Zymogram
Blue Casein Gel
4–16% Tris-Glycine
gel
blue-stained β-
casein
1.5 × 10
–3
units of
trypsin
The following reagents are needed to perform electrophoresis with Novex
™
Zymogram Gels. Ordering information for pre-mixed buffers is on “Accessory
products” on page 80. If you are preparing your own buffers, recipes are provided
on “Recipes” on page 84.
•
Protein sample
•
Deionized water
•
Protein molecular weight markers
•
Novex
™
Tris-Glycine SDS Sample Buffer
•
Novex
™
Tris-Glycine SDS Running Buffer
•
Novex
™
Zymogram Renaturing Buffer
•
Novex
™
Zymogram Developing Buffer
Zymogram
technique
Types of
zymogram gels
Materials
supplied by the
user
Methods
Zymogram gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
27

IMPORTANT!
·
Do not treat zymogram samples with reducing agents. Some proteases are
multiunit complexes that require the full subunit assembly for activity.
·
Load 2–3 times the recommended amount of unstained molecular weight marker
required for a Tris-Glycine Gel. The marker needs to stain intensely to be visualized
against the dark background of the Zymogram Gel.
·
Leave an empty lane between protein molecular weight markers containing
reducing agent and protease sample lanes to prevent diffusion of the reducing
agent into the protease lane.
Use 1X Novex
™
Tris-Glycine SDS Running Buffer for electrophoresis of protease
samples on Zymogram Gels.
1.
Prepare 1,000 mL of Running Buffer as follows:
Reagent Amount
Novex
™
Tris-Glycine SDS Running Buffer
(10X)
100 mL
Deionized Water 900 mL
Total Volume 1,000 mL
2.
Mix thoroughly. Use this buffer to ll the Upper and Lower Buffer Chamber of the
XCell
™
Mini-Cell
™
for electrophoresis.
Prepared samples without reducing agents so that multiunit proteases migrate as a
single unit that can be renatured after electrophoresis.
1.
Prepare each sample as described below:
Reagent Amount
Sample x µL
Novex
™
Tris-Glycine SDS Sample Buffer
(2X)
5 µL
Deionized Water to 5 µL
Total Volume 10 µL
2.
Load the samples onto the gel immediately. Do not heat samples for
Zymogram Gels.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
. Run the gel at 125 V
constant. See “Electrophoresis conditions” on page 47 for additional details on
electrophoresis conditions.
Preparing
running buffer
Preparing
samples
Electrophoresis
conditions
Methods
Zymogram gels
28
Novex
™
Pre-Cast gel electrophoresis guide User Guide
After completing electrophoresis, renature the enzyme and develop the Zymogram
Gels to detect protease activity.
Requirements for the volume of Zymogram Renaturing Buffer and Zymogram
Developing Buffer may vary, depending upon the size of your developing tray.
Up to two mini-gels can be treated with every 100 mL of 1X Novex
™
Zymogram
Renaturing Buffer.
1.
Prepare 100 mL of Renaturing Buffer as described below:
Reagent Amount
Novex
™
Zymogram Renaturing Buffer
(10X)
10 mL
Deionized Water 90 mL
Total Volume 100 mL
2.
Mix thoroughly before use.
Up to two mini-gels can be treated with every 100 mL of 1X Novex
™
Zymogram
Developing Buffer:
1.
Prepare 100 mL of Developing Buffer as described below:
Reagent Amount
Novex
™
Zymogram Developing Buffer
(10X)
10 mL
Deionized Water 90 mL
Total Volume 100 mL
2.
Mix thoroughly before use.
Note: Gels will be treated with Developing Buffer twice, so additional buffer
may be required, depending upon the size of the developing tray.
1.
Remove the gel from the cassette, or remove the top gel plate, and allow the gel
to remain on the bottom gel plate for support.
2.
Incubate the gel in 1X Novex
™
Zymogram Renaturing Buffer for 30 minutes at
room temperature with gentle agitation.
3.
Decant the Zymogram Renaturing Buffer and add 1X Novex
™
Zymogram
Developing Buffer to the gel.
4.
Equilibrate the gel for 30 minutes at room temperature with gentle agitation.
Detecting
protease
activity
Preparing
renaturing
buffer
Preparing
developing
buffer
Developing
zymogram gels
Methods
Zymogram gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
29

5.
Decant the Developing Buffer and add fresh 1X Novex
™
Zymogram Developing
Buffer to the gel.
6.
Incubate the gel at 37℃ for at least 4 hours, or overnight for maximum
sensitivity. The incubation time can be reduced to 1 hour for concentrated
samples. The optimal result is determined empirically by varying the sample load
or incubation time.
Zymogram (Blue Casein) 4–16% gels do not require staining.
For non-pre-stained Zymogram gels, stain the gels with Colloidal Blue Staining Kit or
the SimplyBlue
™
Safestain as described on “SimplyBlue
™
SafeStain
™
protocol” on
page 51–“Colloidal blue staining kit protocol” on page 52.
Areas of protease activity appear as clear bands against a dark background.
Staining
zymogram gels
Methods
Zymogram gels
30
Novex
™
Pre-Cast gel electrophoresis guide User Guide

IEF gels
Isoelectric focusing (IEF) is an electrophoretic technique for the separation of proteins
based on their pI. The pI is the pH at which a protein has no net charge and thus,
does not migrate further in an electric eld.
IEF Gels are used to determine the isoelectric point (pI) of a protein and to detect
minor changes in the protein due to post-translational modications such as
phosphorylation and glycosylation.
In IEF, proteins are applied to polyacrylamide gels (IEF Gels) or immobilized pH
gradient (IPG) strips containing a xed pH gradient. As the protein sample containing
a mixture of proteins migrates through the pH gradient, individual proteins are
immobilized in the pH gradient as they approach their pI.
Novex
™
IEF Gels contain 5% polyacrylamide and are used for native applications.
The pH 3–10 gels have a pI performance range of 3.5–8.5 and the pH 3–7 gels have a
pI performance range of 3.0–7.0.
Proteins separated on IEF Gels are suitable for use in two-dimensional (2D)
electrophoresis using Novex
™
Tris-Glycine or NuPAGE
™
Gels with a 2D-well or
ZOOM
™
format to separate focused proteins by mass.
Two-dimensional (2D) gel electrophoresis is a powerful and sensitive technique for
separating and analyzing protein mixtures from biological samples. 2D gel
electrophoresis is performed in two consecutive steps:
1.
First dimension separation of proteins using isoelectric focusing. Proteins are
separated based on their isoelectric point using IEF gels or IPG strips.
2.
Second dimension separation of proteins using SDS-PAGE.
Proteins are separated based on their molecular weight using denaturing
polyacrylamide gel electrophoresis.
The gel is stained after 2D electrophoresis to visualize the separated proteins, or the
proteins are blotted onto membranes. Protein spots can be excised from the gel or
membranes and subjected to further analyses such as mass spectrometry or
chemical microsequencing to facilitate protein identication.
During IEF, proteins migrate in an electric eld until a stable pH gradient is formed
and the proteins settle into their pI. A high nishing voltage is applied to focus the
proteins into narrow zones. High voltage cannot be used during the initial stages of
IEF as movement of carrier ampholytes generate excessive heat.
To obtain the best results, IEF is typically performed by gradually increasing the
voltage, then maintaining the nal focusing voltage for 30 minutes.
Alternatively, IEF can be performed at constant power, so the voltage will increase as
the current decreases.
Isoelectric
focusing (IEF)
2D
electrophoresis
Power
considerations
for IEF
Methods
IEF gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
31

The following reagents are needed to perform isoelectric focusing with Novex
™
IEF
Gels. Ordering information for pre-mixed buffers is on “Accessory products” on
page 80. If you are preparing your own buffers, recipes are provided on “Recipes”
on page 84.
•
Protein sample
•
Deionized water
•
IEF markers
•
Novex
™
IEF Sample Buffer
•
Novex
™
IEF Cathode Buffer
•
Novex
™
IEF Anode Buffer
•
Fixing solution
Prepare 1X IEF Anode Buffer using Novex
™
IEF Anode Buffer (50X).
1.
Prepare 1,000 mL of IEF Anode Buffer as follows:
Reagent Amount
Novex
™
IEF Anode Buffer (50X) 20 mL
Deionized Water 980 mL
Total Volume 1,000 mL
2.
Mix thoroughly. Use this buffer to ll the Lower Buffer Chamber of the XCell
™
Mini-Cell
™
for electrophoresis.
Prepare 1X IEF Cathode Buffer using the appropriate Novex
™
IEF Cathode Buffer pH
3–10 (10X) or pH 3–7 (10X)
1.
Prepare 200 mL of IEF Cathode Buffer as follows:
Reagent Amount
Novex
™
IEF Cathode Buffer (10X) 20 mL
Deionized Water 180 mL
Total Volume 200 mL
2.
Mix thoroughly. Use this buffer to ll the Upper Buffer Chamber of the XCell
™
Mini-Cell
™
for electrophoresis.
Materials
supplied by the
user
Preparing
anode running
buffer (Lower
buffer
chamber)
Preparing
cathode
running buffer
(Upper buffer
chamber)
Methods
IEF gels
32
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Samples for IEF Gels are prepared without SDS to avoid affecting the pI of the
protein. Reducing agents are also not recommended for the same reason.
1.
Prepare samples for IEF Gels as described below:
Reagent Amount
Sample x µL
Novex
™
IEF Sample Buffer pH 3–10 or
pH 3–7 (2X)
5 µL
Deionized Water to 5 µL
Total Volume 10 µL
2.
Load the sample immediately.
Do not heat samples for IEF Gels.
Fill the Upper Buffer Chamber with
chilled
200 mL 1X IEF Cathode Buffer and the
Lower Buffer Chamber with
chilled
200 mL 1X IEF Anode Buffer.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
. Run the gel at 100 V
constant for 1 hour, followed by 200 V constant for 1 hour, and nish with 500 V
constant for 30 minutes. See “Electrophoresis conditions” on page 47 for additional
details on electrophoresis conditions.
Fixing the proteins in the IEF gel is recommended after electrophoresis. The xing
step also helps to remove carrier ampholytes from the gel, resulting in lower
background after staining.
Fixing solution consists of 12% TCA, or 12% TCA wtih 3.5% sulfosalicylic acid.
1.
Prepare 500 mL of xing solution as follows:
Reagent Amount
Trichloroacetic Acid (TCA) 60.0 g
Sulfosalicylic Acid (optional) 17.5 g
Deionized Water to 500 mL
Total Volume 500 mL
2.
Mix solution thoroughly.
3.
Fix gels for 30 minutes.
IEF gels can be stained by Coomassie
™
or colloidal blue techniques, refer to
“Coomassie
™
staining” on page 50.
If using the SimplyBlue
™
SafeStain
™
, wash the gel extensively to remove traces of
TCA from the xation process to avoid formation of precipitate in the gel.
Preparing
sample
Add anode and
cathode
running buffers
Electrophoresis
conditions
Fixing the gel
Staining IEF
gels
Methods
IEF gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
33

After staining the gel and documenting the results, proteins separated by pI can be
separated by mass.
We recommend using NuPAGE
™
Bis-Tris or Novex
™
Tris-Glycine Gels with a 2D-well,
or ZOOM
™
Gels for 2D SDS-PAGE.
2D-wells can t strips of 6.5 cm, while ZOOM
™
IPG-wells can t strips of 7 cm.
Note: Fixing and staining the IEF gel prior to performing second dimension SDS-
PAGE has the following advantages over other methods of storing IEF gels:
·
Indenite storage without loss of resolution
·
Easy to manipulate as bands are visible
·
Conrms quality of rst dimension IEF before proceeding to SDS-PAGE
In addition to the appropriate gel with a 2D-well or IPG-well, the following reagents
are needed to perform 2D gel electrophoresis with Novex
™
Gels.
•
20% Ethanol
•
Sample Buffer (depending on your gel type)
•
Running Buffer (depending on your gel type)
•
Filter Paper
•
NuPAGE
™
Sample Reducing Agent (optional)
•
Iodoacetamide (optional)
The SDS in the sample buffer and running buffer for SDS-PAGE strips the stain from
proteins and resolubilizes the proteins for migration during 2D electrophoresis.
1.
Incubate the IEF gel in 100 mL 20% ethanol for 10 minutes.
2.
Cut out the desired lane (strip) from the IEF gel for SDS-PAGE.
3.
Incubate the strip in 2 mL 2X SDS sample buffer and 0.5 mL ethanol for 3–
5 minutes. Aspirate the sample buffer and rinse with 1X Running Buffer.
4.
Proceed
Optional Procedure for Reduced Samples:
1.
Incubate the IEF gel in 100 mL 20% ethanol for 10 minutes.
2.
Cut out the desired lane (strip) from the IEF gel for SDS-PAGE.
3.
Incubate the strip in 2 mL 2X SDS sample buffer and 0.5 mL ethanol for 3–
5 minutes. Aspirate the sample buffer and rinse with 1X Running Buffer.
4.
Prepare Reducing Solution by diluting 250 µL of the NuPAGE
™
Sample
Reducing Agent (10X) in 1.75 mL of 1X SDS Sample Buffer.
5.
Incubate the strip in Reducing Solution for 3–5 minutes. Decant the Reducing
Solution.
6.
Prepare 125 mM Alkylating Solution by adding 58 mg of fresh iodoacetamide to
2.5 mL of 1X SDS Sample Buffer.
2D SDS-PAGE
with IEF gels
Materials
supplied by the
user
Equilibrating
the gel
Methods
IEF gels
34
Novex
™
Pre-Cast gel electrophoresis guide User Guide
7.
Incubate the strip in Reducing Solution for 3–5 minutes.
8.
Decant the Alkylating Solution and proceed to 2D Separation of Proteins on
Novex
™
IEF Gels (“2D separation of proteins on Novex
™
IEF gels” on page 35).
A protocol for separating proteins in an IEF gel strip by SDS-PAGE with the XCell
™
Mini-Cell
™
is provided below.
1.
Fill the 2D or IPG-well with the appropriate 1X SDS Running Buffer.
2.
Trim the IEF strip to a length of 5.8–5.9 cm (for 2D-wells) or 6.3–6.4 cm (for
ZOOM
™
IPG-wells) such that the strip includes the pH regions containing your
proteins of interest.
3.
Transfer the IEF gel strip into the well of a 1.0 mm or 1.5 mm gel cassette as
follows:
•
For 1.0 mm Thick Gels
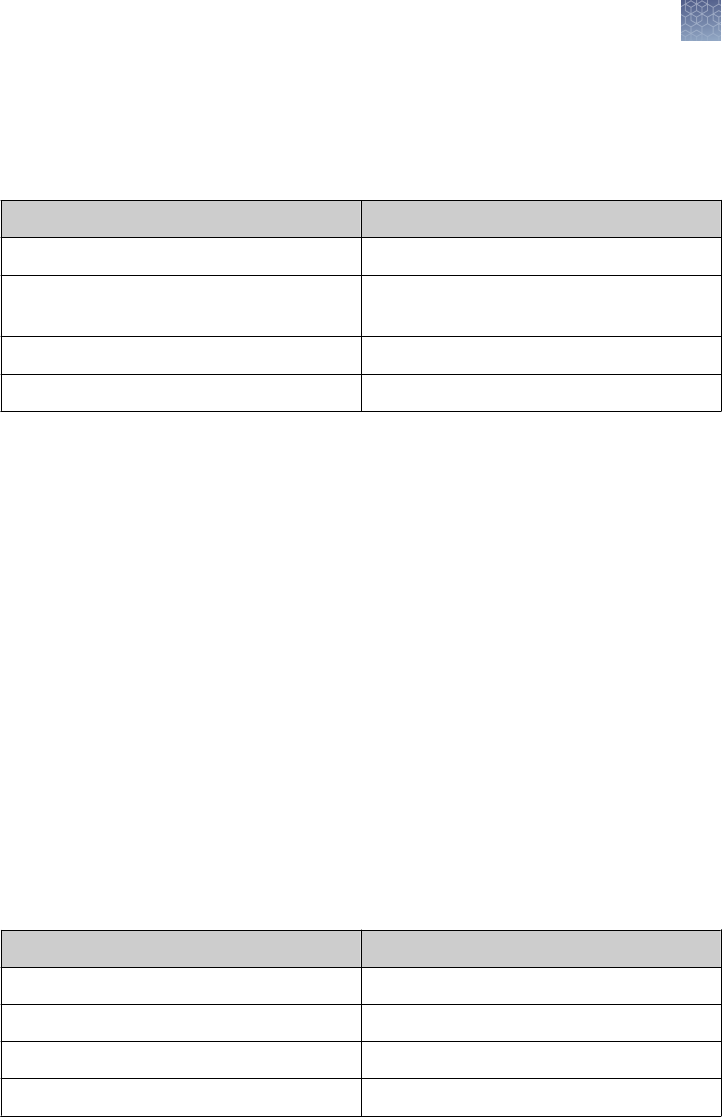
Slide the strip into the well using a gel-loading tip. Avoid trapping air-
bubbles between the gel strip and the surface of the gel. Wet a piece of
thick lter paper (5.8 × 4 cm) in 1X SDS Running Buffer and use it to push
the IEF gel strip down so it makes contact with the surface of the gel (see
gure). The paper should hold the IEF gel strip in place.
•
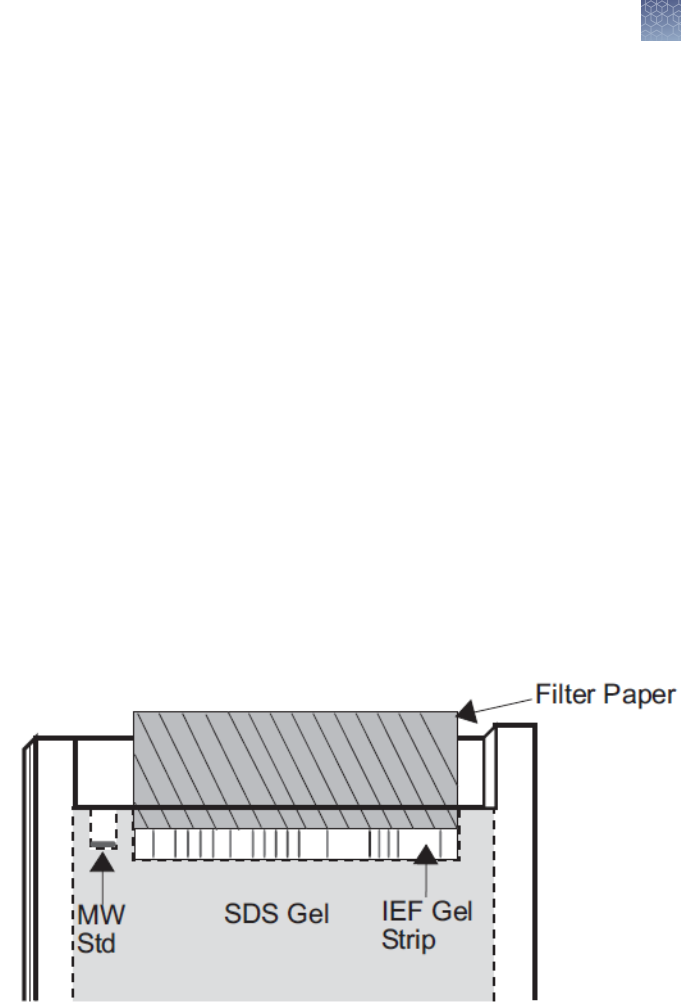
For 1.5 mm Thick Gels
Wet two pieces of thin lter paper (5.8 × 4 cm) in 1X SDS Running Buffer.
Sandwich and the IEF gel strip with the lter paper, such that the edge of
the gel strip protrudes ∼0.5 mm beyond the edge of the paper (see gure).
Insert the sandwich into the well and push the strip so it comes in contact
2D separation
of proteins on
Novex
™
IEF
gels
Methods
IEF gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
35
with the gel. Avoid trapping air-bubbles between the gel strip and the
surface of the gel.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
.
Run the gel at 125 V constant. After the dye front has moved into the stacking gel
(∼10 min), disconnect the power supply, remove the lter paper, and resume
electrophoresis to completion.
Stain the gel with the appropriate method for the type of gel and sample amount after
electrophoresis. Refer to the techniques described on “Coomassie
™
staining” on
page 50–“Using SYPRO
®
Ruby stain as a Post-Stain” on page 62.
Electrophoresis
conditions
Staining the gel
Methods
IEF gels
36
Novex
™
Pre-Cast gel electrophoresis guide User Guide

ZOOM
™
gels
ZOOM
™
Gels are used for 2D analysis of proteins following isoelectric focusing of
IPG strips. ZOOM
™
Gels are 1.0 mm thick, and contain an IPG well and a molecular
weight marker well. The IPG well is designed to accommodate a 7.0 cm IPG strip.
Two types of ZOOM
™
Gels are available (see “Accessory products” on page 80 for
ordering information)
•
NuPAGE
™
Novex
™
4–12% Bis-Tris ZOOM
™
Gel
•
Novex
™
4–20% Tris-Glycine ZOOM
™
Gel
The second dimension electrophoresis procedure involves reducing and alkylating
the proteins focused on your IPG strip in equilibration buffer, loading the strip on your
second dimension gel, and performing SDS-PAGE. For 2D separation of Novex
™
IEF
Gel strips, see “2D SDS-PAGE with IEF gels” on page 34.
You will need the following items for running ZOOM
™
Gels (see “Accessory products”
on page 80 for ordering information):
•
4X NuPAGE
™
LDS Sample Buffer
•
NuPAGE
™
Sample Reducing Agent
•
NuPAGE
™
Novex
™
4–12% Bis-Tris ZOOM
™
Gel or Novex
™
4–20% Tris-Glycine
ZOOM
™
Gel
•
Running Buffer (depending on your gel type)
•
0.5% agarose solution
•
Iodoacetamide
•
Plastic exible ruler or thin weighing spatula
•
15 mL conical tubes
•
Water bath set at 55℃ or 65℃
•
Protein molecular weight marker
1.
Dilute 4X NuPAGE
™
LDS Sample Buffer to 1X with deionized water.
2.
Add 500 µL of the NuPAGE
™
Sample Reducing Agent (10X) to 4.5 mL of the 1X
NuPAGE
™
LDS Sample Buffer from Step 1 on page 37 in a 15 mL conical tube.
Place one IPG strip in this conical tube for equilibration.
3.
Incubate for 15 minutes at room temperature. Decant the Reducing Solution.
4.
Prepare 125 mM Alkylating Solution by adding 116 mg of fresh iodoacetamide
to 5 mL of 1X NuPAGE
™
LDS Sample Buffer from Step 1 on page 37.
5.
Add 5 mL of Alkylating Solution (from Step 4 on page 37) to the conical tube
containing the IPG strip. Incubate for 15 minutes at room temperature.
6.
Decant the Alkylating Solution and proceed immediately to SDS-PAGE, “SDS-
PAGE” on page 38.
ZOOM
™
gels
2D separation
of IPG strips
Materials
supplied by the
user
Equilibrating
the IPG strip
Methods
ZOOM
™
gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
37

A protocol for separating proteins in an IPG strip by SDS-PAGE with ZOOM
™
Gels
and the XCell
™
Mini-Cell
™
is provided below.
1.
Prepare 0.5% agarose solution in the appropriate running buffer and keep it
warm (55–65℃) until you are ready to use the agarose solution.
2.
Cut the plastic ends of the IPG strip ush with the gel. Do not cut off any
portions of the gel.
3.
Slide the IPG strip into the ZOOM
™
Gel well.
4.
If the molecular weight marker well is bent, straighten the well using a gel-
loading tip.
5.
Align the IPG strip properly in the ZOOM
™
Gel well using a thin plastic ruler or a
weighing spatula. Avoid trapping air bubbles between the strip and the gel while
sliding the strip into the well.
6.
Pour ∼ 400 µL of 0.5% agarose solution into the ZOOM
™
Gel well to seal the
IPG strip in place. Make sure the agarose solution does not overow into the
molecular weight marker well.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 for instructions on
running Novex
™
Pre-Cast Gels using the XCell
™
Mini-Cell
™
.
Note: Do not use the ZOOM
™
IPGRunner
™
Core
™
for electrophoresis of the second
dimension gel. You must use the Buffer Core
™
supplied with the XCell
™
Mini-Cell
™
.
Perform electrophoresis at 200 V for 40 minutes for NuPAGE
™
Novex
™
Bis-Tris
ZOOM
™
Gels or at 125 V for 90 minutes for Novex
™
Tris-Glycine ZOOM
™
Gels.
Stain the gel with the appropriate method for the type of gel and sample amount after
electrophoresis. Refer to the techniques described on “Coomassie
™
staining” on
page 50–“Using SYPRO
®
Ruby stain as a Post-Stain” on page 62.
SDS-PAGE
Electrophoresis
conditions
Staining the gel
Methods
ZOOM
™
gels
38
Novex
™
Pre-Cast gel electrophoresis guide User Guide

TBE gels
Novex
™
polyacrylamide TBE Gels provide high-resolution analysis of restriction
digests and PCR products. The TBE Gels give sharp, intense bands and provide
separations of double-strand DNA fragments from 10–3,000 base pairs.
Using polyacrylamide gels for nucleic acid separation provides the following
advantages over agarose gels:
•
High resolution and sensitivity
•
Lower background staining
•
Requires less sample concentration and volume
•
Efcient blotting
•
Easy to extract DNA from the gel and does not interfere with enzymatic reactions
•
Accurate and reproducible results
The following reagents are needed to perform gel electrophoresis with Novex
™
TBE
Gels. Ordering information for pre-mixed buffers is on “Accessory products” on
page 80. If you are preparing your own buffers, recipes are provided on “Recipes”
on page 84.
•
DNA sample
•
Deionized water
•
Appropriate DNA markers
•
Novex
™
Hi-Density TBE Sample Buffer
•
Novex
™
TBE Running Buffer
Use 1X Novex
™
TBE Running Buffer to perform electrophoresis.
1.
Prepare 1,000 mL of Running Buffer as follows:
Reagent Amount
Novex
™
TBE Running Buffer (5X) 200 mL
Deionized Water 800 mL
Total Volume 1,000 mL
2.
Mix thoroughly. Use this buffer to ll the Upper and Lower Buffer Chamber of the
XCell
™
Mini-Cell
™
for electrophoresis.
Introduction
Advantages of
TBE gels
Materials
supplied by the
user
Preparing
running buffer
Methods
TBE gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
39

Novex
™
TBE Gels require only ∼10% of the amount of sample used on large gels or
agarose gels. Dilute your standards and samples to ∼ 0.01 OD (0.2 µg/band) to avoid
overloading the gel.
1.
Prepare samples for TBE gels as described below:
Reagent Amount
Sample x µL
Novex
™
Hi-Density TBE Sample Buffer
(5X)
2 µL
Deionized Water to 8 µL
Total Volume 10 µL
2.
Load the samples immediately on the gel.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 instructions for running
TBE Gel using the XCell
™
Mini-Cell
™
. Run the gel at 200 V constant. See
“Electrophoresis conditions” on page 47 for additional details on electrophoresis
conditions.
The size of the DNA fragments visualized at the dye fronts of the different TBE Gels is
shown in the table below.
Gel Type
Dye Front
[1]
Bromophenol Blue (dark
blue)
Xylene Cyanol (blue green)
6% TBE Gel 65 bp 250 bp
8% TBE Gel 25 bp 220 bp
10% TBE Gel 35 bp 120 bp
20% TBE Gel 15 bp 50 bp
4–12% TBE Gel 35 bp 400 bp
4–20% TBE Gel 25 bp 300 bp
[1]
accuracy is ± 5 bp
Novex
™
TBE Gels can be stained by silver staining, ethidium bromide, and SYBR
™
Green staining techniques after electrophoresis. Refer to “Silver staining” on
page 54–“Ethidium bromide staining” on page 64
Preparing
samples
Electrophoresis
conditions
Migration of
the dye fronts
Staining the gel
Methods
TBE gels
40
Novex
™
Pre-Cast gel electrophoresis guide User Guide

TBE-Urea gels
Novex
™
denaturing polyacrylamide TBE-Urea Gels provide high resolution of short
single-strand oligonucleotides. The gels provide excellent resolution for fast size and
purity conrmations of DNA or RNA oligos from 20–600 bases.
The TBE-Urea Gels contain 7 M urea for maximum denaturation.
The following reagents are needed to perform gel electrophoresis with Novex
™
TBE-
Urea Gels. Ordering information for pre-mixed buffers is on “Accessory products” on
page 80. If you are preparing your own buffers, recipes are provided on “Recipes”
on page 84.
•
DNA or RNA sample
•
Deionized water
•
Appropriate DNA or RNA markers
•
Novex
™
TBE-Urea Sample Buffer
•
Novex
™
Prep TBE-Urea Sample Buffer (for preparative electrophoresis only)
•
Novex
™
TBE Running Buffer
Note: To obtain optimal results with TBE-Urea Gels, observe the following
recommendations:
·
Use RNase-free ultrapure water
·
Prior to loading samples, ush wells several times with 1X TBE Running Buffer to
remove urea
·
Load samples quickly and avoid allowing the gel to stand for long periods of time
after loading to prevent diffusion
·
Use Prep TBE-Urea Sample Buffer for preparative gel electrophoresis as this buffer
does not contain any marker dyes
·
Wear gloves and use dedicated equipment to prevent contamination
·
Avoid using buffers with formamide on TBE-Urea polyacylamide gels as it will
result in fuzzy bands
Use 1X Novex
™
TBE Running Buffer to perform electrophoresis.
1.
Prepare 1,000 mL of Running Buffer as follows:
2.
Reagent Amount
Novex
™
TBE Running Buffer (5X) 200 mL
Deionized Water 800 mL
Total Volume 1,000 mL
3.
Mix thoroughly. Use this buffer to ll the Upper and Lower Buffer Chamber of the
XCell
™
Mini-Cell
™
for electrophoresis.
4.
Flush wells of the gel several times with 1X TBE Running Buffer to remove urea
from the wells prior to loading samples to obtain sharp bands.
Introduction
Materials
supplied by the
user
Preparing
running buffer
Methods
TBE-Urea gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
41

Novex
™
TBE-Urea Gels require only ∼10% of the amount of sample used on large
gels or agarose gels. Dilute your standards and samples to ∼ 0.01 OD (0.2 µg/band)
to avoid overloading the gel.
1.
Prepare samples for TBE-Urea Gels as described below:
Reagent Amount
Sample x µL
Novex
™
TBE-Urea Sample Buffer (2X) 5 µL
Deionized Water to 5 µL
Total Volume 10 µL
2.
Heat samples at 70℃ for 3 minutes to denature the samples.
3.
Load the samples immediately on the gel. If the samples are not used
immediately, place them on ice to prevent renaturation.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 instructions for running
TBE-Urea Gel using the XCell
™
Mini-Cell
™
. Run the gel at 180 V constant. See
“Electrophoresis conditions” on page 47 for additional details on electrophoresis
conditions.
The size of the single-strand DNA fragments visualized at the dye fronts of the
different TBE-Urea Gels is shown in the table below.
Gel Type
Dye Front
[1]
Bromophenol Blue (dark
blue)
Xylene Cyanol (light blue)
6% TBE-Urea Gel 25 bases 110 bases
10% TBE-Urea Gel 20 bases 55 bases
15% TBE-Urea Gel 10 bases 40 bases
[1]
accuracy is ± 5 bases
Novex
™
TBE-Urea Gels can be stained by silver staining, ethidium bromide, and
SYBR
™
Green staining techniques after electrophoresis. Refer to “Coomassie
™
staining” on page 50–“Ethidium bromide staining” on page 64 for more information
on these techniques.
Preparing
samples
Electrophoresis
conditions
Migration of
the dye fronts
Staining the gel
Methods
TBE-Urea gels
42
Novex
™
Pre-Cast gel electrophoresis guide User Guide

DNA retardation gels
Novex
™
DNA Retardation Gels consist of 6% Polyacrylamide prepared with 0.5X TBE
as the gel buffer. The 6% gel provides good resolution of fragments in the range of
60–2500 bp used for DNA retardation assays.
The gel shift assay is based on the fact that the movement of a DNA molecule
through a non-denaturing Polyacrylamide gel is hindered when bound to a protein
molecule (Revzin, 1989). This technique is used to characterize DNA/protein
complexes. The 0.5X TBE buffer is sufcient for good electrophoretic separation yet
low enough to promote DNA/ protein interactions.
Detection is performed with ethidium bromide staining of DNA or, for greater
sensitivity, with radiolabeling the DNA or protein.
The following reagents are needed to perform gel electrophoresis with Novex
™
DNA
Retardation Gels. Ordering information for pre-mixed buffers is on “Accessory
products” on page 80. If you are preparing your own buffers, recipes are provided
on “Recipes” on page 84.
•
DNA sample
•
Deionized water
•
Novex
™
Hi-Density TBE Sample Buffer
•
Novex
™
TBE Running Buffer
1.
Prepare samples for DNA Retardation Gels as described below:
Reagent Amount
Sample x µL
Novex
™
Hi-Density TBE Sample Buffer
(5X)
1 µL
Deionized Water to 9 µL
Total Volume 10 µL
2.
Load the samples immediately on the gel.
Note: Specic buffer conditions may be required during incubation of the protein
and DNA target sequence in order to minimize non-specic DNA/protein interactions
for certain samples.
If salt concentration is low (0.1 M or less), the samples can usually be loaded in the
incubation buffer after adding about 3–5% glycerol and a small amount of
bromophenol blue tracking dye.
Gel-Shift assay
Materials
supplied by the
user
Preparing
samples
Methods
DNA retardation gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
43
Prepare 1,000 mL of 0.5X Novex
™
TBE Running Buffer as follows:.
1.
Reagent Amount
Novex
™
TBE Running Buffer (5X) 100 mL
Deionized Water 900 mL
Total Volume 1,000 mL
2.
Mix thoroughly. Use this buffer to ll the Upper and Lower Buffer Chamber of the
XCell
™
Mini-Cell
™
for electrophoresis.
See “Electrophoresis of Novex
™
Pre-Cast gels” on page 45 instructions for running
DNA Retardation Gels using the XCell
™
Mini-Cell
™
. Run the gel at 100 V constant.
See “Electrophoresis conditions” on page 47 for additional details on
electrophoresis conditions.
Gel-shift assays use labeled (radioactive, uorescent, biotin) DNA fragments for
visualization of results. Use the appropriate technique to develop the image for the
type of label you are using.
Preparing
running buffer
Electrophoresis
conditions
Staining the gel
Methods
DNA retardation gels
44
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Electrophoresis of Novex
™
Pre-Cast gels
Instructions are provided below for electrophoresis of Novex
™
Pre-Cast Gels using
the XCell
™
Mini-Cell
™
. For more information on the XCell
™
Mini-Cell
™
, refer to the
manual (IM-9003). This manual is available on our website or contact Technical
Support.
For information on sample and buffer preparation for Novex
™
Pre-Cast Gels, see
“General guidelines for preparing samples and buffers” on page 20–“Preparing
running buffer” on page 44.
Wear gloves and safety glasses when handling gels.
XCell
™
Mini-Cell
™
requires 200 mL for the Upper Buffer Chamber and 600 mL for the
Lower Buffer Chamber.
1.
Remove the Novex
™
Pre-Cast Gel from the pouch.
2.
Rinse the gel cassette with deionized water. Peel off the tape from the bottom of
the cassette.
3.
Gently pull the comb out of the cassette in one smooth motion.
4.
Rinse the sample wells with the appropriate 1X Running Buffer. Invert the gel
and shake the gel to remove the buffer. Repeat two more times.
5.
Orient the two gels in the Mini-Cell
™
such that the notched “well” side of the
cassette faces inwards toward the Buffer Core
™
. Seat the gels on the bottom of
the Mini-Cell
™
and lock into place with the Gel Tension Wedge. Refer to the
XCell
™
Mini-Cell
™
manual (IM-9003) for detailed instructions.
Note: If you are running just one gel, use the plastic Buffer Dam in place of the
second gel cassette to form the Upper Buffer Chamber.
6.
Fill the Upper Buffer Chamber with a small amount of the Running Buffer to
check for tightness of seal. If you detect a leak from Upper to the Lower Buffer
Chamber, discard the buffer, reseal the chamber, and check the seal again.
7.
Once the seal is tight, ll the Upper Buffer Chamber (Inner) with the appropriate
1X Running Buffer. The buffer level must exceed the level of the wells.
8.
Load an appropriate volume of sample at the desired protein concentration onto
the gel (see “Recommended loading volumes” on page 18 for recommended
loading volumes).
9.
Load appropriate protein molecular weight markers (see “Protein stains and
standards” on page 82 for ordering information).
10.
Fill the Lower Buffer Chamber with 600 mL of the appropriate 1X Running Buffer.
Introduction
Protocol using
XCell
™
Mini-
Cell
™
Methods
Electrophoresis of Novex
™
Pre-Cast gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
45

11.
Place the XCell
™
Mini-Cell
™
lid on the Buffer Core
™
. With the power on the
power supply turned off, connect the electrode cords to the power supply [red
to (+) jack, black to (–) jack].
12.
See “Electrophoresis conditions” on page 47
Methods
Electrophoresis of Novex
™
Pre-Cast gels
46
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Power supply settings for Novex
™
Pre-Cast gels
Run your gels according to the following protocol:
Gel Type Voltage Expected Current
[1]
Run Time
Tris-Glycine Gels
(SDS-PAGE)
125 V constant Start: 30–40mA
End: 8–12 mA
90 minutes
(dependent on gel
type)
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
Tris-Glycine Gels
(Native-PAGE)
125 V constant Start: 6–12 mA
End: 3–6 mA
1–12 hours
Tricine Gels 125 V constant Start: 80 mA
End: 40 mA
90 minutes
(dependent on gel
type)
Run the gel until the
phenol red tracking
dye reaches the
bottom of the gel.
Zymogram Gels 125 V constant Start: 30–40 mA
End: 8–12 mA
90 minutes
(dependent on gel
type)
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
IEF Gels 100 V constant:
1 hour
200 V constant:
1 hour
500 V constant:
30 min
Start: 5 mA
End: 6 mA
2.5 hours
Electrophoresis
conditions
Methods
Power supply settings for Novex
™
Pre-Cast gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
47
Gel Type Voltage Expected Current
[1]
Run Time
TBE Gels 200 V constant
[2]
Start: 10–18 mA
End:4–6 mA
30–90 minutes
(dependent on gel
type)
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
6% TBE-Urea
Gels
180 V constant
[2]
Start: 19 mA
End: 14 mA
50 minutes
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
10% TBE-Urea
Gels
180 V constant
[2]
Start: 15 mA
End: 8 mA
60 minutes
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
15% TBE-Urea
Gels
180 V constant
[2]
Start: 13 mA
End: 6 mA
75 minutes
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
DNA
Retardation Gels
100 V constant Start: 12–15 mA
End: 6–15 mA
90 minutes
Run the gel until the
bromophenol blue
tracking dye reaches
the bottom of the
gel.
[1]
Expected start and end current values are stated for single gels.
[2]
Voltages up to 250 V may be used to reduce the run time.
Methods
Power supply settings for Novex
™
Pre-Cast gels
48
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Opening Novex
™
Pre-Cast gel cassettes
1.
After electrophoresis is complete, shut off the power, disconnect electrodes, and
remove gel(s) from the XCell
™
Mini-Cell
™
.
2.
Separate each of the three bonded sides of the cassette by inserting the Gel
Knife into the gap between the two plastic plates that make up the cassette. The
notched (“well”) side of the cassette should face up.
3.
Push down gently on the knife handle to separate the plates. Repeat on each
side of the cassette until the plates are completely separated.
CAUTION! Use caution while inserting the Gel Knife between the two plates to
avoid excessive pressure on the gel.
4.
Carefully remove and discard the top plate, allowing the gel to rest on the
bottom (slotted) plate.
5.
If blotting, proceed to “Blotting Novex
™
Pre-Cast gels” on page 68 without
removing the gel from the bottom plate.
6.
If staining, remove the gel from the plate by one of the methods:
•
Use the sharp edge of the Gel Knife to remove the gel foot from the bottom
of the gel. Hold the Gel Knife at a 90° angle, perpendicular to the gel and
the slotted half of the cassette. Push down on the knife, and then repeat the
motion across the gel to cut off the entire foot. Hold the plate and gel over a
container with the gel facing downward and use the knife to carefully loosen
one lower corner of the gel and allow the gel to peel away from the plate.
•
Hold the plate and gel over a container with the gel facing downward.
Gently push the Gel Knife through the slot in the cassette, until the gel peels
away from the plate. Cut the gel foot off of the gel after xing and staining,
but before drying.
7.
Fix and stain the gel as described on “Coomassie
™
staining” on page 50–
“Ethidium bromide staining” on page 64. For developing the Zymogram gel for
enzyme activity, see “Preparing running buffer” on page 28. For xing IEF gels,
see “Fixing the gel” on page 33.
Removing the
gel after
electrophoresis
Methods
Opening Novex
™
Pre-Cast gel cassettes
Novex
™
Pre-Cast gel electrophoresis guide User Guide
49

Coomassie
™
staining
Instructions are provided below for Coomassie
™
staining Tris-Glycine, Zymogram,
IEF, and Tricine Gels using the SimplyBlue
™
SafeStain
™
, Colloidal Blue Staining Kit,
and Coomassie
™
R-250.
If you are using other types of Coomassie
™
staining kits, follow the appropriate
manufacturer′s recommendations.
Note: If you are staining low molecular weight peptides (< 2.5 kDa), we recommend
xing the gel in 5% glutaraldehyde and 50% methanol for one hour and then follow
the instructions in the Colloidal Blue Staining Kit Manual (IM-6025) for small peptides.
Guidelines and apparent molecular weight values for Novex
™
protein molecular
weight standards are provided on “Assigned apparent molecular weights” on
page 76.
You will need the following items for staining your gel (see “Accessory products” on
page 80 for ordering information):
•
Staining container
•
Deionized water
•
Orbital Shaker
For SimplyBlue
™
SafeStain
™
(see “SimplyBlue
™
SafeStain
™
protocol” on page 51):
•
SimplyBlue
™
SafeStain
™
•
Optional: 20% NaCl
•
Optional: Microwave oven
•
12% Trichloroacetic acid (for IEF gels)
For Colloidal Blue Staining Kit (see “Colloidal blue staining kit protocol” on
page 52):
•
Colloidal Blue Staining Kit
•
Methanol
•
Optional: 20% Ammonium sulfate
•
Fixing solutino (for IEF gels)
For Coomassie
™
R-250 Staining (see “Coomassie
™
R-250 staining protocol” on
page 52):
•
0.1% Coomassie
™
R-250 in 40% ethanol and 10% acetic acid
•
Destaining Solution consisting of 10% ethanol and 7.5% acetic acid
•
Optional: Microwave oven
Introduction
Molecular
weight
calibration
Materials
supplied by the
user
Methods
Coomassie
™
staining
50
Novex
™
Pre-Cast gel electrophoresis guide User Guide

The Basic Protocol for staining Novex
™
Gels with SimplyBlue
™
SafeStain
™
is
provided below. For the Microwave Protocol and staining large format gels, refer to
the SimplyBlue
™
SafeStain
™
Manual (IM-6050). This manual is available on our
website or contact Technical Support.
For general use with 1.0 mm and 1.5 mm thick Tris-Glycine Gels, and 1.0 mm thick
Tricine, Zymogram, and IEF Gels (8 cm × 8 cm).
After electrophoresis follow the instructions below. Be sure the mini-gel moves freely
in water or stain to facilitate diffusion during all steps.
Note: Stain Zymogram Gels with SimplyBlue
™
SafeStain
™
after renaturing and
developing the gel for enzyme activity.
1.
Fix IEF Gels in 100 mL 12% TCA for 15 minutes. The xing step is not required
for Tris-Glycine, Tricine, and Zymogram Gels.
2.
Rinse the mini-gel 3 times for 5 minutes with 100 mL deionized water to remove
SDS and buffer salts, which interfere with binding of the dye to the protein.
Discard each rinse.
3.
Stain the mini-gel with enough SimplyBlue
™
SafeStain
™
(20–100 mL) to cover
the gel. Stain for 1 hour at room temperature with gentle shaking. Bands will
begin to develop within minutes. After incubation, discard the stain. Stain cannot
be re-used.
Note: Gel can be stained for up to 3 hours, but after 3 hours, sensitivity will
decrease. If you need to leave the gel overnight in the stain, add 2 mL of 20%
NaCl (w/v) in water for every 20 mL of stain. This procedure will not affect
sensitivity.
4.
Wash the mini-gel with 100 mL of water for 1–3 hours. The gel can be left in the
water for several days without loss of sensitivity. There is a small amount of dye
in the water that is in equilibrium with the dye bound to the protein, so proteins
will remain blue.
5.
To obtain the clearest background for photography, perform a second 1 hour
wash with 100 mL water.
Note: Sensitivity decreases at this point if the gel is allowed to stay in the
water more than 1 day. Reduction of free dye in the water favors dissociation
of the dye from the protein. If you need to store the gel in water for a few days,
add 20 mL of 20% NaCl.
6.
For gel drying, see “Gel drying” on page 65.
SimplyBlue
™
SafeStain
™
protocol
Methods
Coomassie
™
staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
51

A brief staining protocol for staining Novex
™
Gels with the Colloidal Blue Staining Kit
is provided below. For more details on the staining procedure, refer to the Manual
(IM-6025). This manual is available on our website or contact Technical Support.
1.
Fix the IEF Gel in xing solution as described on “Fixing the gel” on page 33.
This step is not required for Tris-Glycine, Tricine, and Zymogram Gels.
2.
Prepare staining solution for a single gel as described in the table below. For two
gels, double the volume of reagents used for staining. Be sure to shake Stainer
B prior to making the solution.
Solutions
Tris-Glycine, Tricine, and
Zymogram Gel
IEF Gel
Deionized Water 55 mL 58 mL
Methanol 20 mL 20 mL
Stainer B 5 mL 2 mL
Stainer A 20 mL 20 mL
3.
Incubate the gel in this staining solution as follows at room temperature with
gentle shaking:
•
Tris-Glycine, Tricine, and Zymogram Gels for a minimum of 3 hours and a
maximum of 12 hours.
•
IEF Gels for 30 minutes.
4.
Decant staining solution and add a minimum of 200 mL of deionized water per
gel to the staining container. Gently shake gel in water for at least 7 hours. Gel
will have a clear background after 7 hours in water.
5.
For gel drying, see “Gel drying” on page 65.
Note: Novex
™
Gels can be left in deionized water for up to 3 days without signicant
change in band intensity and background clarity.
For long-term storage (over 3 days), keep the gel in a 20% ammonium sulfate
solution at 4℃.
The Coomassie
™
staining protocol described below is recommended for staining
Novex
™
Gels. You may use any Coomassie
™
staining protocol of choice.
1.
Prepare the staining solution containing 0.1% Coomassie
™
R-250 in 40%
ethanol, 10% acetic acid.
2.
After electrophoresis, incubate 1 or 2 gels in a staining container containing
100 mL Coomassie
™
Blue R-250 staining solution.
CAUTION! Use caution while performing the following steps using a microwave
oven. Do not overheat the staining solutions.
Colloidal blue
staining kit
protocol
Coomassie
™
R-250 staining
protocol
Methods
Coomassie
™
staining
52
Novex
™
Pre-Cast gel electrophoresis guide User Guide

3.
Loosely cover the staining container and heat in a microwave oven at full power
for 1 minute. To prevent hazardous, ammable vapors from forming, do not
allow the solution to boil.
4.
Remove the staining container from the microwave oven and gently shake the
gel for 15 minutes at room temperature on an orbital shaker.
5.
Decant the stain and rinse the gel once with deionized water.
6.
Prepare a destain solution containing 10% ethanol and 7.5% acetic acid.
7.
Place one or two stained gels in a staining container containing the 100 mL
destain solution.
8.
Loosely cover the staining container and heat in a microwave oven at full power
for 1 minute.
9.
Gently shake the gel at room temperature on an orbital shaker until the desired
background is achieved.
10.
For gel drying, see “Gel drying” on page 65.
Methods
Coomassie
™
staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
53

Silver staining
Instructions are provided below for silver staining Novex
™
Gels using the
SilverQuest
™
Silver Staining Kit and the SilverXpress
™
Silver Staining Kit (see
“Accessory products” on page 80 for ordering information).
If you are using any other silver staining kit, follow the manufacturer′s
recommendations.
Guidelines and apparent molecular weight values for Novex
™
protein molecular
weight standards are provided on “Protein stains and standards” on page 82.
You will need following items for silver staining your gel (see “Accessory products” on
page 80 for ordering information):
•
Staining container
•
Rotary Shaker
•
Ultrapure water (>18 megohm/cm resistance recommended)
•
Teon
™
coated stir bars
•
Disposable 10 mL pipettes
•
Clean glass bottles for reagent preparation
•
Graduated glass cylinders
•
Protein molecular weight markers (Mark12
™
Unstained Standard, recommended)
For SilverQuest
™
Staining:
•
SilverQuest
™
Silver Staining Kit
•
30% ethanol (made with ultrapure water)
•
100% ethanol
•
Fixative (40% ethanol, 10% acetic acid, made with ultrapure water)
For SilverXpress
™
Staining:
•
SilverXpress
™
Silver Staining Kit
•
Methanol
•
Acetic acid
•
Sulfosalicylic acid
•
Trichloroacetic acid (TCA)
Introduction
Molecular
weight
calibration
Materials
supplied by the
user
Methods
Silver staining
54
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Note: For optimal silver staining results, follow these guidelines:
·
Be sure to wear clean gloves that have been rinsed with deionized water while
handling gels
·
Use clean containers and designate these containers for silver staining purposes
only
·
Make sure the size of the container permits free movement of the gel during
shaking and complete immersion in solution while staining
·
Do not touch the gel with bare hands or metal objects and do not put pressure on
gels while handling or changing solutions
·
Use teon coated stir bars and clean glass containers to prepare reagents
·
Avoid cross contamination of kit reagents
·
Use freshly made solutions
Use the reagents provided in the SilverQuest
™
Silver Staining Kit to prepare the
following solutions for staining:
•
Sensitizing solution
Ethanol 30 mL
Sensitizer 10 mL
Ultrapure water to 100 mL
•
Staining solution
Stainer 1 mL
Ultrapure water to 100 mL
•
Developing solution
Developer 10 mL
Developer enhancer 1 drop
Ultrapure water to 100 mL
Note: You may prepare all solutions immediately before starting the staining protocol
or prepare them as you proceed to the next step.
Preparing
solutions for
SilverQuest
™
silver staining
Methods
Silver staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
55

SilverQuest
™
microwave silver staining protocol
The Fast Staining protocol (using a microwave oven) for silver staining Novex
™
Gels
using SilverQuest
™
Silver Staining Kit is described below. For the Basic Protocol and
more details on the staining procedure, refer to the SilverQuest
™
Silver Staining Kit
Manual (IM-6070). This manual is available on our website or contact Technical
Support.
Use 100 mL of each solution for each 1.0 mm thick, 8 × 8 cm Novex
™
Gel.
Note: You may have to optimize the staining protocol, if the dimensions of your gel
are not the same as mentioned above.
CAUTION! Use caution while performing the Fast Staining Protocol using a
microwave oven. Do not overheat the staining solutions.
1.
After electrophoresis, place the gel in a clean microwaveable staining tray of the
appropriate size. Rinse the gel briey with ultrapure water.
2.
Place the gel in 100 mL of xative and microwave at high power (700 watts) for
30 seconds. Remove the gel from the microwave and gently agitate it for
5 minutes at room temperature. Decant the xative.
3.
Wash the gel with 100 mL of 30% ethanol in a microwave at high power for
30 seconds. Remove the gel from the microwave and gently agitate it for
5 minutes at room temperature on a rotary shaker. Decant the ethanol.
4.
Add 100 mL of Sensitizing solution to the washed gel. Microwave at high power
for 30 seconds. Remove the gel from the microwave and place it on a rotary
shaker for 2 minutes at room temperature. Decant the Sensitizing solution.
5.
Add 100 mL ultrapure water to the gel. Microwave at high power for 30 seconds.
Remove the gel from the microwave and gently agitate it for 2 minutes at room
temperature. Decant the water, and repeat the step one more time.
6.
Place the gel in 100 mL of Staining solution. Microwave at high power for
30 seconds. Remove the gel from the microwave and gently agitate it for
5 minutes at room temperature.
7.
Decant the Staining solution and wash the gel with 100 mL of ultrapure water for
20–60 seconds. Do not wash the gel for more than a minute.
8.
Place the gel in 100 mL of Developing solution and incubate for 5 minutes at
room temperature with gentle agitation on a rotary shaker. Do not microwave.
9.
Once the desired band intensity is achieved, immediately add 10 mL of Stopper
directly to the gel still immersed in Developing solution and gently agitate the gel
for 10 minutes. The color changes from pink to clear indicating the end of
development.
10.
Wash the gel with 100 mL of ultrapure water for 10 minutes. For gel drying, see
“Gel drying” on page 65.
Methods
Silver staining
56
Novex
™
Pre-Cast gel electrophoresis guide User Guide
If you need to destain the gel for mass spectrometry analysis, see the SilverQuest
™
Silver Staining Kit Manual (IM-6070).
Prepare the reagents as described below. If you are staining two gels, double the
reagent volumes.
•
Fixing solution for Tris-Glycine and Tricine Gels
Methanol 100 mL
Acetic Acid 20 mL
Ultrapure water to 200 mL
•
Fixing solution
for TBE, TBE-Urea Gels
Sulphosalicylic acid 7 g
TCA 24 g
Ultrapure water to 200 mL
•
Sensitizing solution
Methanol 100 mL
Sensitizer 5 mL
Ultrapure water to 200 mL
•
Staining solution
Stainer A 5 mL
Stainer B 5 mL
Ultrapure water 90 mL
•
Developing Solution
Developer 5 mL
Ultrapure water 95 mL
Preparing
solutions for
SilverXpress
™
silver staining
Methods
Silver staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
57
The following staining procedure is for 1 mm thick Novex
™
Gels. If you are using
1.5 mm thick Novex
™
Gels, double the incubation time.
For gel drying, see “Gel drying” on page 65.
Note: Gels may be stored in the second Sensitizing Solution overnight, if desired.
Step Solution Vol/Gel
Gel Type
Tris-Glycine Tricine TBE/TBE-Urea IEF
1A Fix the gel in
Fixing
Solution.
200 mL 10 minutes 10 minutes 10 minutes 10 minutes
1B N/A N/A N/A N/A 10 minutes
2A Decant the
Fixing Solution
and incubate
the gel in two
changes of
Sensitizing
Solution.
100 mL 10 minutes 30 minutes 10 minutes 30 minutes
2B 100 mL 10 minutes 30 minutes 10 minutes 30 minutes
3A Decant the
Sensitizing
Solution and
rinse the gel
twice with
ultrapure
water.
200 mL 5 minutes 5 minutes 5 minutes 5 minutes
3B 200 mL 5 minutes 5 minutes 5 minutes 5 minutes
4 Incubate the
gel in Staining
Solution.
100 mL 15 minutes 15 minutes 30 minutes 15 minutes
5A Decant the
Staining
Solution and
rinse the gel
twice with
ultrapure
water.
200 mL 5 minutes 5 minutes 5 minutes 5 minutes
5B 200 mL 5 minutes 5 minutes 5 minutes 5 minutes
6 Incubate the
gel in
Developing
Solution.
100 mL 3–15 minutes 3–15 minutes 3–15 minutes 3–15 minutes
SilverXpress
™
silver staining
protocol
Methods
Silver staining
58
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Step Solution Vol/Gel
Gel Type
Tris-Glycine Tricine TBE/TBE-Urea IEF
7 Add the
Stopping
Solution
directly to the
gel when the
desired
staining
intensity is
reached.
5 mL 10 minutes 10 minutes 10 minutes 10 minutes
8A Decant the
Stopping
Solution and
wash the gel
three times in
ultrapure
water.
200 mL 10 minutes 10 minutes 10 minutes 10 minutes
8B 200 mL 10 minutes 10 minutes 10 minutes 10 minutes
8C 200 mL 10 minutes 10 minutes 10 minutes 10 minutes
Methods
Silver staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
59

SYPRO
®
Ruby staining
Instructions are provided below for a basic and rapid protocol for Novex
™
Pre-Cast
Gels (Novex
™
Tris-glycine gels, Novex
™
Tricine gels, ZOOM
™
gels, and Novex
™
IEF
gels) for the detection of proteins, including glycoproteins and phosphoproteins.
SYPRO
®
Ruby provides the following advantages:
•
Linear quantitation range of over three orders of magnitude
•
Compatible with subsequent analysis of proteins by Edman based sequencing or
mass spectrometry in 1D or 2D format
•
Compatible with non-denaturing gels and IEF gels (basic protocol)
Guidelines and apparent molecular weight values for Novex
™
protein molecular
weight standards are provided on “Protein stains and standards” on page 82.
You will need following items for silver staining your gel (see “Accessory products” on
page 80 for ordering information):
•
SYPRO
®
Ruby gel stain
•
Staining containers, 1 per gel (see below for details)
•
Reagent-grade methanol
•
Reagent-grade glacial acetic acid
•
Trichloroacetic acid (for IEF gels only)
•
Ultrapure water (18 megohm-cm recommended)
•
Rotary shaker
•
Powder-free latex or vinyl gloves
•
Microwave oven (700–1200 W) (optional)
•
Water bath set at 80℃ (optional)
Note: General considerations for the protocol include the following:
·
Perform all xation, staining, and washing steps with continuous, gentle agitation
(e.g., on an orbital shaker at 50 rpm).
·
We recommend polypropylene or polycarbonate containers for staining. Glass
dishes are not recommended. Staining containers should be meticulously clean to
minimize contamination and other artifacts.
·
For convenience, gels may be left in x solution overnight or longer.
·
For convenience, gels may be left in SYPRO
®
Ruby stain indenitely without
overstaining, although speckling artifacts tend to increase over time.
·
As with any uorescent stain, cover the gel container during staining and
subsequent wash steps to exclude light.
Introduction
Advantages of
SYPRO
®
Ruby
staining
Molecular
weight
calibration
Materials
supplied by the
user
Methods
SYPRO
®
Ruby staining
60
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Prepare the reagents as described below. If you are staining two gels, double the
reagent volumes. Increase volumes 1.5‑fold for 1.5mm thick gels.
•
Fix Solution
Methanol 100 mL
Glacial Acetic Acid 14 mL
Ultrapure water to 200 mL
•
Fix Solution for IEF Gels
Methanol 40 mL
Trichloroacetic Acid 10 g
Ultrapure water to 100 mL
•
Wash Solution
Methanol 10 mL
Glacial Acetic Acid 7 mL
Ultrapure water to 100 mL
The basic protocol results in the maximum signal strength and widest linear dynamic
range for staining of denaturing gels, non-denaturing gels, and IEF gels. Sensitivity is
in the 1 ng range for most proteins.
1.
After electrophoresis, place the gel into a clean container with 100 mL of Fix
Solution and agitate on an orbital shaker for 30 minutes. Pour off the used x
solution and repeat once more with fresh Fix Solution.
Note: For IEF Gels, place the gel into a clean container with 100 mL of IEF Fix
Solution and agitate on an orbital shaker for 3 hours. After xing, perform 3
washes in ultrapure water for 10 minutes each, before proceeding to the staining
step.
2.
Pour off the used x solution.
3.
Add 60 mL of SYPRO
®
Ruby gel stain to the tray containing the gel. Agitate on
an orbital shaker overnight.
4.
Transfer the gel to a clean container and wash in 100 mL of Wash Solution for
30 minutes. The transfer step helps minimize background staining irregularities
and stain speckles on the gel.
5.
Rinse the gel in ultrapure water for 5 minutes. Repeat the rinse a minimum of
one more time to prevent possible corrosive damage to your imager.
Note: If you are staining two gels, double the reagent volumes. Increase volumes
1.5‑fold for 1.5mm thick gels.
Preparing
solutions for
SYPRO
®
Ruby
staining
SYPRO
®
Ruby
basic protocol
Methods
SYPRO
®
Ruby staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
61

Proteins stained with SYPRO
®
Ruby protein gel stain are readily visualized using a UV
or blue-light source. The use of a photographic camera or CCD camera and the
appropriate lters is essential to obtain the greatest sensitivity.
SYPRO
®
Ruby stain can be used to post-stain gels stained with other gel stains such
as Pro-Q
™
Diamond phosphoprotein gel stain, Pro-Q
™
Emerald 300 glycoprotein gel
stain, Pro-Q
™
Sapphire
™
or InVision
™
oligohistidine-tag gel stains, or Pro-Q
™
Amber
transmembrane protein gel stain.
Always use SYPRO
®
Ruby stain last, as the SYPRO
®
Ruby signal can dominate the
signal from other stains. SYPRO
®
Ruby stain does not work well as a post-stain for
colorimetric stains such as Coomassie
™
and silver stains.
Visualization of
SYPRO
®
Ruby
stained gels
Using SYPRO
®
Ruby stain as a
Post-Stain
Methods
SYPRO
®
Ruby staining
62
Novex
™
Pre-Cast gel electrophoresis guide User Guide

SYBR
™
green staining
The SYBR
™
Green I nucleic acid gel stain is a sensitive stain that can be used to
detect DNA in Novex
™
TBE and TBE-Urea Gels. As little as 20–60 pg of double
stranded DNA, 100–300 pg of single stranded DNA, or 1–2 ng of a synthetic 24-mer
can be detected, depending upon the wavelength of transillumination.
Note: General considerations for the protocol include the following:
·
We recommend polypropylene containers for staining. Glass dishes are not
recommended. Staining containers should be meticulously clean to minimize
contamination and other artifacts.
·
SYBR
™
Green I reagent has optimal sensitivity at pH 7.5–8.0.
·
For convenience, gels may be left in SYBR
™
Green I stain for up to 24 hours with
little decrease in sensitivity.
Perform post-staining of DNA on TBE or TBE-Urea Gels as follows:
1.
Prepare a 1:10,000 dilution of SYBR
™
Green I reagent in TE (10 mM Tris-HCl,
1 mM EDTA, pH 8.0), TBE, or TAE buffer.
2.
Remove the gel from the cassette using a Gel Knife, and place it in a
polypropylene staining container.
3.
Cover the gel with staining solution and incubate at room temperature for 10–
40 minutes with gentle agitation. Protect the staining container from light by
covering it with aluminum foil.
SYBR
™
Green I stain is compatible with a wide variety of gel reading instruments,
ranging from ultraviolet transilluminators to argon laser and mercury-arc lamp
excitation gel scanners. SYBR
™
Green I stain is maximally excited at 497 nm, but
also has secondary excitation peaks at ∼290 nm and ∼380 nm. The uorescence
emission of SYBR
™
Green I stain bound to DNA is centered at 520 nm.
Introduction
Procedure
Visualization of
SYBR
™
green I
stained gels
Methods
SYBR
™
green staining
Novex
™
Pre-Cast gel electrophoresis guide User Guide
63

Ethidium bromide staining
A brief protocol is provided below for staining nucleic acids on TBE and TBE-Urea
Gels with ethidium bromide.
Procedure
CAUTION! Ethidium bromide is a powerful mutagen and is moderately toxic. Wear
gloves and protective clothing when handling ethidium bromide solutions.
1.
Prepare 2 µg/ml solution of ethidium bromide in ultrapure water.
2.
Remove the gel from the cassette using a Gel Knife, and place it in a staining
container.
3.
Incubate the gel in the ethidium bromide solution for 20 minutes.
4.
Destain the gel by rinsing the gel three times with ultrapure water for 10 minutes.
Note: Ethidium bromide staining of polyacrylamide gels requires at least 10 ng of
DNA for detection due to the quenching of the uorescence by polyacrylamide.
For alternative techniques with greater detection sensitivity, perform silver staining
using the SilverXpress
™
Silver Staining Kit (see “Preparing solutions for SilverXpress
™
silver staining” on page 57) or SYBR
™
Green I staining (see “SYBR
™
green staining”
on page 63).
Introduction
Methods
Ethidium bromide staining
64
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Gel drying
Dry gels by passive evaporation (air-drying) or vacuum drying. Vacuum drying is
faster than passive air-drying methods but often results in cracked gels due to the
speed of dehydration.
We recommend drying Novex
™
Pre-Cast gels using passive air-drying methods such
as the DryEase
™
Mini-Gel Drying System (see below). For applications that require
vacuum drying, follow the recommendations on “Vacuum drying” on page 66 to
minimize cracking of the gels.
You will need the following items for drying your gel (see “Accessory products” on
page 80 for ordering information):
•
DryEase
™
Mini-Gel Drying System
•
Gel-Dry
™
Drying Solution (or prepare your own gel drying solution containing
30% methanol and 5% glycerol)
•
StainEase
™
Gel Staining Tray or a suitable round container
Silver stained and Coomassie
™
stained Novex
™
Gels can be dried by vacuum drying
or by air-drying. We recommend using the DryEase
™
Mini-Gel Drying System to air-
dry the gel.
A brief gel drying protocol using the DryEase
™
Mini-Gel Drying System is provided
below. For more details on this system, refer to the DryEase
™
Mini-Gel Drying System
manual (IM-2380). This manual is available for download from our website or contact
Technical Support.
1.
After all staining and destaining steps are complete, wash the destained gel(s)
three times for two minutes each time in deionized water (50 mL per mini-gel) on
a rotary shaker.
2.
Decant the water and add fresh Gel-Dry
™
Drying Solution (35 mL per minigel).
3.
Equilibrate the gel in the Gel-Dry
™
Drying Solution by shaking the gel for 15–
20 minutes in the StainEase
™
Gel Staining Tray or in a round container.
Note: Do not equilibrate gels stained with Coomassie
™
G-250 in the Gel-Dry
™
Drying Solution for more than 5 minutes to avoid losing band intensity.
4.
Cut any rough edges off the gel (including the wells and the gel foot) using the
Gel Knife or a razor blade.
5.
Remove 2 pieces (per gel) of cellophane from the package.
6.
Immerse one sheet at a time in the Gel-Dry
™
Drying Solution. Allow 10 seconds
for complete wetting before adding additional sheets. Do not soak the
cellophane for more than 2 minutes.
7.
Place one side of the DryEase
™
Gel Drying Frame with the corner pin facing up,
on the DryEase
™
Gel Drying Base.
Introduction
Materials
supplied by the
user
DryEase
™
Mini-
Gel drying
system
Methods
Gel drying
Novex
™
Pre-Cast gel electrophoresis guide User Guide
65

8.
Center a piece of pre-wetted cellophane from Step 5 on page 65 over the
base/frame combination, so the cellophane lays over the inner edge of the
frame.
9.
Lay the gel on the center of the cellophane sheet making sure no bubbles are
trapped between the gel and the cellophane. Add some Gel-Dry
™
Drying
Solution to the surface of the cellophane, if necessary.
10.
Carefully lay the second sheet of cellophane over the gel so that no bubbles are
trapped between the cellophane and the gel. Add some Gel-Dry
™
Drying
Solution if necessary. Gently smooth out any wrinkles in the assembly with a
gloved hand.
11.
Align the remaining frame so that its corner pins t into the appropriate holes on
the bottom frame. Push the plastic clamps onto the four edges of the frames.
12.
Lift the frame assembly from the DryEase
™
Gel Drying Base and pour off the
excess solution from the base.
13.
Stand the gel dryer assembly upright on a bench top. Be careful to avoid drafts
as they can cause an uneven rate of dying which leads to cracking. Drying takes
between 12–36 hours depending on humidity and gel thickness.
14.
When the cellophane is dry to touch, remove the gel/cellophane sandwich from
the drying frame. Trim off the excess cellophane.
15.
Press the dried gel(s) between the pages of a notebook under light pressure for
approximately 2 days so they remain at for scanning, photography, display, and
overhead projection.
General guidelines are provided below to minimize cracking during vacuum drying of
gels. For detailed instructions on vacuum drying, follow the manufacturer′s
recommendations.
Handle Gels with Care:
Remove the gel from the cassette without breaking or tearing the edges. Small nicks
or tears can act as a starting point for cracking. Remove the gel wells and foot off the
bottom of the gel with a Gel Knife or a razor blade as described on “Opening Novex
™
Pre-Cast gel cassettes” on page 49. Use the StainEase
™
Staining Tray for staining
and destaining gels. This tray is designed to facilitate the solution changing process
without handling of gels.
Use a Gel Drying Solution:
We recommend equilibrating the gel in a gel drying solution such as Gel-Dry
™
Gel
Drying Solution for 10–30 minutes at room temperature with gentle shaking on an
orbital shaker before drying the gel. Gel-Dry
™
Gel Drying Solution contains a
proprietary non-glycerol component to effectively regulate the rate of drying and
prevent cracking. The gel drying solutions do not interfere with autoradiography.
To prepare your own gel drying solution, prepare a solution containing 30% methanol
and 5% glycerol.
Vacuum drying
Methods
Gel drying
66
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Note: Do not incubate gels stained with Coomassie
™
G-250 in gel drying solution for
more than 5 minutes as the bands may fade.
Remove Air Bubbles:
Remove any air bubbles that may be trapped between the paper, gel, and plastic
wrap by rolling a small glass pipette over the gel. Use additional gel drying solution to
help remove the air bubbles.
Use Proper Gel Dryer Set-up:
Place gel on the gel dryer with the plastic wrap facing up. Make sure the vacuum
pump is in working condition, and properly set up to form a tight seal when on. Use
drying conditions for polyacrylamide gels, with the temperature increasing to a set
value and holding for the duration of the drying cycle. We recommend drying mini-
gels at 80℃ for 2 hours.
Ensure Gel is Completely Dry:
The gel will crack if the vacuum seal of the heated gel dryer is broken prior to
complete drying of the gel. To ensure the gel is completely dried before releasing the
vacuum seal, follow these tips :
•
Check the temperature of the gel
The temperature of the dried gel should be the same as the temperature of the
surrounding gel drying surface. If the temperature of the dried gel is cooler, then
the gel is not completely dried.
•
Check for moisture in the tubing connecting the gel dryer to the vacuum pump
The gel is not completely dried if there is residual moisture in the tubing and
additional drying time is required.
Methods
Gel drying
Novex
™
Pre-Cast gel electrophoresis guide User Guide
67

Blotting Novex
™
Pre-Cast gels
After performing electrophoresis, proteins can be transferred to membranes for
subsequent analysis. Methods of transfer include wet, semi-wet, semi-dry, and dry
blotting. Semi-dry blotting can be performed with the Novex
™
Semi-Dry Blotter, and
dry blotting is performed with the iBlot
™
Gel Transfer Device. Refer to the respective
manuals for information on blotting with these devices.
Instructions are provided below for semi-wet blotting of Novex
™
Pre-Cast Gels using
the XCell II Blot Module. For more information on the XCell II Blot Module, refer to the
manual (IM-9051).
If you are using any other blotting apparatus, follow the manufacturer′s
recommendations.
During blotting, the distance traveled (gel thickness) between the electrodes is much
lower than during electrophoresis requiring lower voltage and lower eld strength
(volts/distance). However, the cross sectional area of current ow is much greater
requiring higher current.
Blotting power requirements depend on eld strength (electrode size) and
conductivity of transfer buffer. The higher the eld strength and conductivity of the
buffer, the higher is the current requirement (the current decreases during the run as
the ions in the buffer polarize). It is important to use a power supply capable of
accommodating the initial high current requirement.
In addition to the XCell II Blot Module, the following reagents are needed for blotting
your gel (see “Accessory products” on page 80 for ordering information):
•
Blotting membranes
•
Filter paper (not needed if using Novex
™
pre-cut membrane/lter paper
sandwiches)
•
Methanol (if using PVDF membranes)
•
Appropriate Transfer Buffer
•
Deionized water
Introduction
Power
considerations
for blotting
Materials
supplied by the
user
Methods
Blotting Novex
™
Pre-Cast gels
68
Novex
™
Pre-Cast gel electrophoresis guide User Guide
For blotting Tris-Glycine, Tricine, and IEF Gels use 1X Tris-Glycine Transfer Buffer. If
you are preparing your own transfer buffer see “Tris-Glycine transfer buffer” on
page 86 for a recipe.
An alternate transfer protocol for IEF Gels is provided on “Blotting IEF gels” on
page 74.
If you are performing protein sequencing, an alternate transfer buffer compatible with
the technique is listed on the .
Prepare 1,000 mL of Transfer Buffer:
Tris-Glycine Transfer Buffer (25X) 40 mL
Methanol 200 mL
Deionized Water 760 mL
Total Volume 1,000 mL
For blotting TBE and TBE-Urea Gels, use 0.5X TBE Running Buffer. If you are
preparing your own transfer buffer, see “TBE running buffer” on page 89 for a
recipe.
Prepare 1,000 mL of 1X Tris-Glycine Transfer Buffer using the Tris-Glycine Transfer
Buffer (25X) as follows:
TBE Running Buffer (5X)
40 mL
Methanol 200 mL
Deionized Water 760 mL
Total Volume 1,000 mL
For blotting TBE and TBE-Urea Gels
Dilute the 5X TBE Running Buffer to 0.5X with deionized water.
Tris-Glycine Transfer Buffer interferes with protein sequencing. If you are performing
protein sequencing, use the NuPAGE
™
Transfer Buffer or the 0.5X TBE Running
Buffer to perform blotting.
The NuPAGE
™
Transfer Buffer protects against modication of the amino acid side
chains and is compatible with N-terminal protein sequencing using Edman
degradation.
Use about 700 mL of 1X Transfer Buffer to soak the pads until saturated. Remove the
air bubbles by squeezing the pads while they are submerged in buffer. Removing the
air bubbles is essential as they can block the transfer of biomolecules if they are not
removed.
Preparing
transfer buffer
Preparing
transfer buffer
for TBE gels
Preparing
transfer buffer
compatible
with protein
sequencing
Preparing
blotting pads
Methods
Blotting Novex
™
Pre-Cast gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
69

Cut the transfer membrane and lter paper to the dimensions of the gel or use
Novex
™
pre-cut membrane/lter paper sandwiches.
•
PVDF membrane—Pre-wet PVDF membrane for 30 seconds in methanol,
ethanol, or isopropanol. Briey rinse in deionized water, then place in a shallow
dish with 50 mL of 1X Transfer Buffer for several minutes.
•
Nitrocellulose—Place the membrane directly into a shallow dish containing
50 mL of 1X Transfer Buffer for several minutes.
•
Filter paper—Soak the lter paper briey in 1X Transfer Buffer immediately prior
to use.
•
Gel
—Use the gel immediately following the run.
Do not soak the gel in transfer
buffer.
Wear gloves while performing the blotting procedure to prevent contamination of gels
and membranes, and exposure to irritants commonly used in electrotransfer.
Transferring One Gel
1.
After opening the gel cassette as described on “Opening Novex
™
Pre-Cast gel
cassettes” on page 49, remove wells with the Gel Knife.
2.
Place a piece of pre-soaked lter paper on top of the gel, with the edge above
the slot in the bottom of the cassette (leaving the foot of the gel uncovered).
Keep the lter paper saturated with the transfer buffer and remove all trapped air
bubbles by gently rolling over the surface using a glass pipette as a roller.
3.
Turn the plate over so the gel and lter paper are facing downwards over a
gloved hand or clean at surface.
4.
Use the Gel Knife to push the foot out of the slot in the plate, and separate the
gel from the plate.
5.
When the gel is on a at surface, cut the foot off the gel with the Gel Knife.
6.
Wet the surface of the gel with transfer buffer and position the pre-soaked
transfer membrane on the gel, ensuring all air bubbles have been removed.
7.
Place another pre-soaked lter paper on top of the membrane. Remove any
trapped air bubbles.
Preparing
transfer
membrane and
filter paper
Western
transfer using
the XCell II blot
module
Methods
Blotting Novex
™
Pre-Cast gels
70
Novex
™
Pre-Cast gel electrophoresis guide User Guide
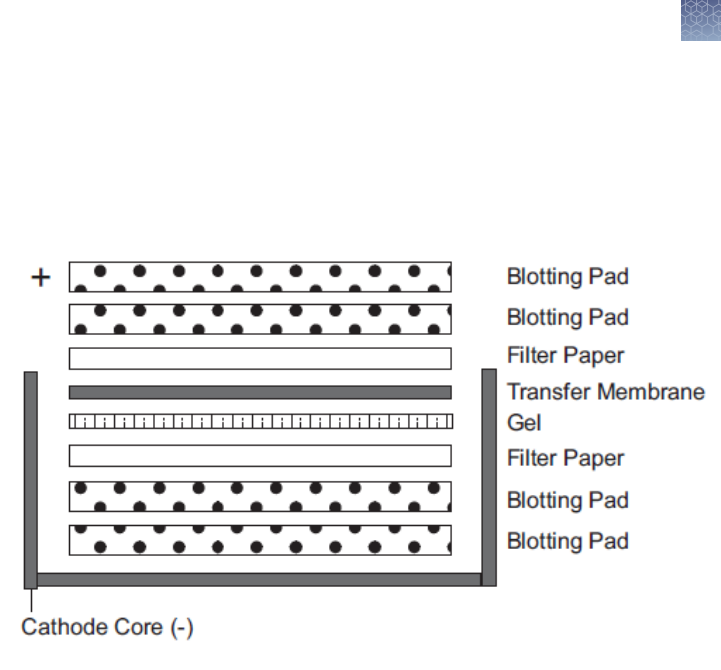
8.
Place two soaked blotting pads into the cathode (–) core of the blot module. The
cathode core is the deeper of the two cores and the corresponding electrode
plate is a darker shade of gray. Carefully pick up the gel/membrane assembly
and place it on the pad such that the gel is closest to the cathode plate (see
Figure 1).
Figure 1
9.
Add enough pre-soaked blotting pads to raise the assembly 0.5 cm over the
edge of cathode core. Place the anode (+) core on top of the pads. The
gel/membrane assembly should be held securely between the two halves of the
blot module ensuring complete contact of all components.
10.
Position the gel membrane sandwich and blotting pads in the cathode core of
the XCell II Blot Module to t horizontally across the bottom of the unit. There
should be a gap of approximately 1 cm at the top of the electrodes when the
pads and assembly are in place.
11.
Hold the blot module together rmly and slide it into the guide rails on the Lower
Buffer Chamber. The blot module ts into the unit only one way, with the (+) sign
at the upper left hand corner of the blot module, and the inverted gold post
tting into the connector on the right side of the Lower Buffer Chamber.
12.
Place the gel tension wedge so that its vertical face is against the blot module.
Lock the gel tension wedge by pulling the lever forward.
13.
Fill the blot module with 1X Transfer Buffer until the gel/membrane sandwich is
covered in Transfer Buffer. To avoid generating extra conductivity and heat, do
not ll the chamber all the way to the top.
Methods
Blotting Novex
™
Pre-Cast gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
71
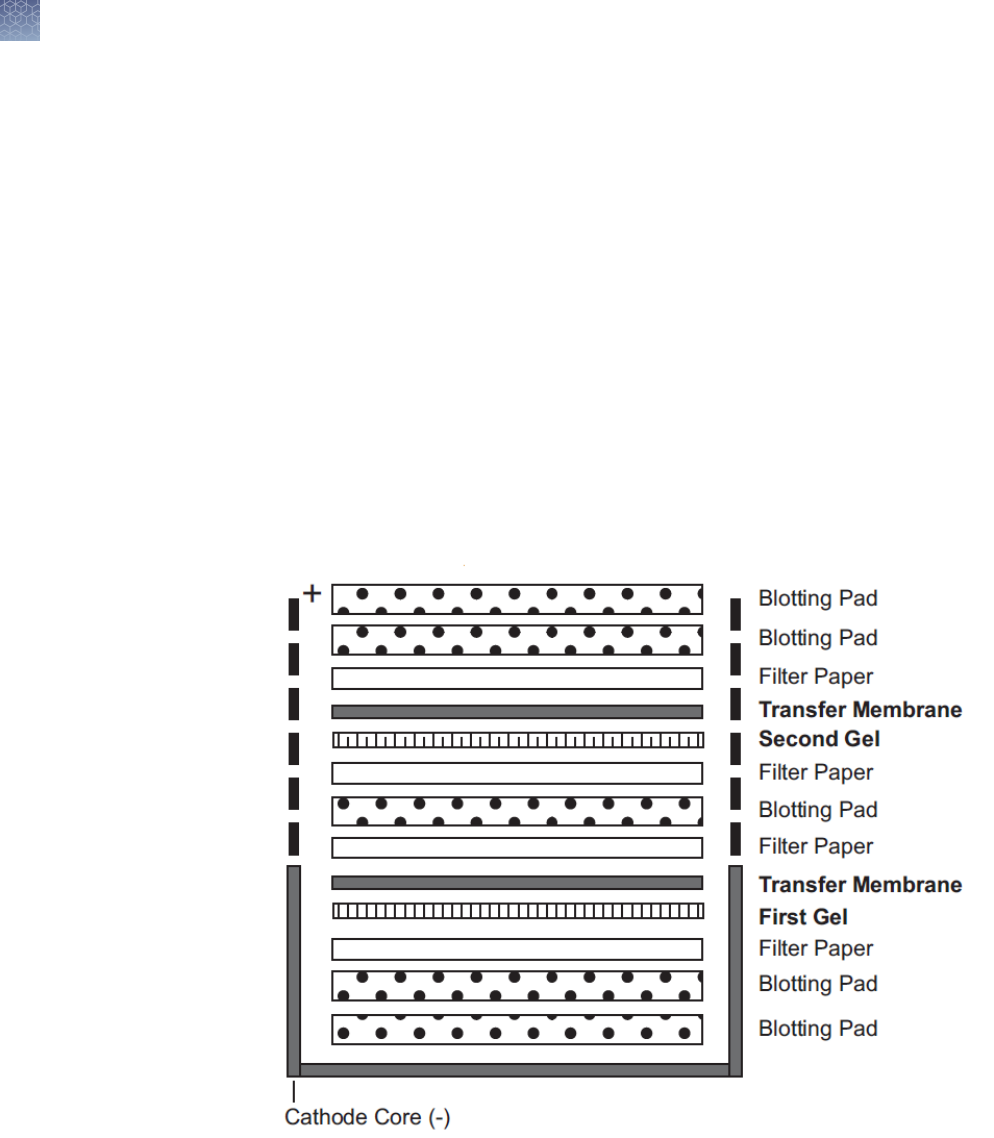
14.
Fill the Lower Buffer Chamber with deionized water by pouring approximately
650 mL in the gap between the front of the blot module and the front of the
Lower Buffer Chamber. The water level should reach approximately 2 cm from
the top of the Lower Buffer Chamber. This serves to dissipate heat produced
during the run.
15.
Place the lid on top of the unit.
16.
With the power turned off, plug the red and black leads into the power supply.
Refer to “Recommended transfer conditions” on page 73 for transfer
conditions.
Transferring Two Gels in One Blot Module
1.
Repeat Steps 1–7 on page 70 () twice to make two gel-membrane assemblies.
2.
Place two pre-soaked pads on cathode shell of blot module. Place the rst
gel/membrane assembly on the pads such that the gel faces the cathode plate.
(See Figure 2).
Figure 2
3.
Add another pre-soaked blotting pad on top of rst gel/membrane assembly.
4.
Position second gel/membrane assembly on top of blotting pad with the gel
facing the cathode side.
5.
Proceed with steps 8–13 on page 71 from Transferring One Gel.
6.
Refer to “Recommended transfer conditions” on page 73 for transfer
conditions.
Methods
Blotting Novex
™
Pre-Cast gels
72
Novex
™
Pre-Cast gel electrophoresis guide User Guide
The transfer conditions for Novex
™
Pre-Cast Gels using the XCell II Blot Module are
listed in the table below.
Note: The expected current listed in the table is for transferring one gel. If you are
transferring two gels in the blot module, the expected current is roughly twice the
listed value.
Gel Transfer Buffer Membrane Power Conditions
Tris-Glycine Gel
Tricine Gel
1X Tris-Glycine
Transfer Buffer with
20% methanol
Nitrocellulose or
PVDF
25 V constant for 1–
2 hours
Expected Current
Start: 100 mA
IEF Gel 1X Tris-Glycine
Transfer Buffer with
20% methanol
Nitrocellulose or
PVDF
25 V constant for
1 hour
Expected Current
Start: 65–85 mA
0.7% Acetic acid pH
3.0
See for details on
this alternate
transfer protocol.
Nitrocellulose or
PVDF
10 V constant for
1 hour
Expected Current
Start: 65–85 mA
TBE Gel 0.5X TBE Running
Buffer
Nylon 30 V constant for
1 hour
Expected Current
Start: 39 mA
End: 35 mA
TBE-Urea Gel 0.5X TBE Running
Buffer
Nylon 30 V constant for
1 hour
Expected Current
Start: 39 mA
End: 35 mA
DNA Retardation
Gel
0.5X TBE Running
Buffer
Nylon 30 V constant for
1 hour
Expected Current
Start: 39 mA
End: 35 mA
Recommended
transfer
conditions
Methods
Blotting Novex
™
Pre-Cast gels
Novex
™
Pre-Cast gel electrophoresis guide User Guide
73

Novex
™
IEF Gels are composed of 5% polyacrylamide and are more susceptible to
hydrolysis due to the heat generated with the recommended blotting protocol. The
following protocol has been optimized to prevent hydrolysis and effective transfer of
basic proteins due to the low pH of the transfer buffer.
1.
Prepare chilled 0.7% acetic acid.
2.
After electrophoresis, remove the gel from the cassette and equilibrate the gel in
the 0.7% acetic acid for 10 minutes.
STOPPING POINT The 5% polyacrylamide gels are stickier and more difcult to
handle than higher percentage polyacrylamide gels. To prevent the gel from
sticking to the ler paper before it is in the proper position, remove the gel from
the equilibration solution by submerging a piece of lter paper under the gel
while it is oating in the equilibration solution. When the gel and lter paper are
in the correct position, lift the lter paper so that it attaches to the gel.
3.
Assemble the gel/membrane sandwich as described on “Western transfer using
the XCell II blot module” on page 70, except in
reverse order
so that the
membrane is on the cathode (–) side of the gel.
4.
Transfer for 1 hour at 10 V constant.
During SDS-PAGE all proteins have a net negative charge due to the SDS in the
sample buffer and the running buffer. Proteins separated during native gel
electrophoresis do not have a net charge which may cause problems during the
transfer. Some native proteins may have a higher pI than the pH of the Tris-Glycine
Transfer Buffer used in standard transfer protocols. Guidelines are provided below to
increase the transfer efciency of native proteins.
•
Increasing the pH of the transfer buffer to 9.2 (25 mM Tris Base, 25 mM glycine,
pH 9.2), allows proteins with pI below 9.2 to transfer towards the anode
electrode
•
Place a membrane on both sides of the gel if you are using the regular Tris-
Glycine Transfer Buffer, pH 8.3. If there are any proteins that are more basic than
the pH of the transfer buffer, they will be captured on the membrane placed on
the cathode side of the gel
•
Incubate the gel in 0.1% SDS for 15 minutes before blotting with Tris-Glycine
Transfer Buffer. The small amount of SDS will render enough charge to the
proteins so they can move unidirectionally towards the anode and in most cases
will not denature the protein
Native proteins may diffuse out of the membrane into the solution during the blocking
or antibody incubation steps, as the native proteins tend to be more soluble. To
prevent diffusion of the proteins out of the membrane, we recommend xing the
proteins to the membrane by air drying the membrane or incubating the membrane in
5–10% acetic acid for 15 minutes followed by rinsing the membrane with deionized
water and then air drying.
By performing any of these two xing methods the proteins will be sufciently
unfolded to expose hydrophobic sites and bind more efciently to the membrane.
Blotting IEF
gels
Blotting native
gels
Methods
Blotting Novex
™
Pre-Cast gels
74
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Calibrating protein molecular weight
The molecular weight of a protein can be determined based upon its relative mobility
by constructing a standard curve with protein standards of known molecular weights.
The protein mobility in SDS-PAGE gels is dependent on the
•
Length of the protein in its fully denatured state,
•
SDS-PAGE buffer systems
•
Secondary structure of the protein
An identical molecular weight standard may have slightly different mobility resulting in
different apparent molecular weight when run in different SDS-PAGE buffer systems.
If you are using the Novex
™
protein molecular weight standards, see the apparent
molecular weights of these standards on the Novex
™
Pre-Cast Gels listed on the to
determine an apparent molecular weight of your protein.
When using SDS-PAGE for molecular weight determination, slight deviations from the
calculated molecular weight of a protein (calculated from the known amino acid
sequence) can occur due to the retention of varying degrees of secondary structure
in the protein, even in the presence of SDS. This phenomenon is observed in highly
organized secondary structures (such as collagens, histones, or highly hydrophobic
membrane proteins) and in peptides, where the effect of local secondary structure
and amino acid side chains becomes magnied relative to the total size of the
peptide.
Slight differences in protein mobilities also occur when the same proteins are run in
different SDS-PAGE buffer systems. Each SDS-PAGE buffer system has a different
pH, which affects the charge of a protein and its binding capacity for SDS. The
degree of change in protein mobility is usually small in natural proteins but more
pronounced with “atypical” or chemically modied proteins such as pre-stained
standards.
Introduction
Protein
secondary
structure
Buffer systems
Methods
Calibrating protein molecular weight
Novex
™
Pre-Cast gel electrophoresis guide User Guide
75
Values for apparent molecular weight of Novex
™
molecular weight standards are
derived from the construction of a calibration curve in the Tris-Glycine SDS-PAGE
System. We have now calculated and assigned apparent molecular weights for the
Novex
™
protein standards in several buffer systems. Remember to use the one that
matches your gel for the most accurate calibration of your protein.
The following charts summarize the approximate molecular weight values for the
Novex
™
protein molecular weight standards when run in different buffer systems. You
may generate calibration curves in your lab with any other manufacturer′s standards.
Novex
™
Sharp Pre-stained
Protein Standard
Tris-Glycine Gels (4–20%) Tricine Gels (10–20%)
Band 1 260 kDa 260 kDa
Band 2 160 kDa 160 kDa
Band 3 110 kDa 110 kDa
Band 4 80 kDa 80 kDa
Band 5 60 kDa 60 kDa
Band 6 50 kDa 50 kDa
Band 7 40 kDa 40 kDa
Band 8 30 kDa 30 kDa
Band 9 20 kDa 20 kDa
Band 10 15 kDa 15 kDa
Band 11 10 kDa 10 kDa
Band 12
3.5 kDa
Mark12
™
Unstained
Standard
Tris-Glycine Gels (4–20%) Tricine Gels (10–20%)
Myosin 200 kDa 200 kDa
β-Galactosidase 116.3 kDa 116.3 kDa
Phosphorylase B 97.4 kDa 97.4 kDa
Bovine Serum Albumin 66.3 kDa 66.3 kDa
Glutamic Dehydrogenase 55.4 kDa 55.4 kDa
Lactate Dehydrogenase 36.5 kDa 36.5 kDa
Carbonic Anhydrase 31 kDa 31 kDa
Trypsin Inhibitor 21.5 kDa 21.5 kDa
Lysozyme 14.4 kDa 14.4 kDa
Assigned
apparent
molecular
weights
Methods
Calibrating protein molecular weight
76
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Mark12
™
Unstained
Standard
Tris-Glycine Gels (4–20%) Tricine Gels (10–20%)
Aprotinin 6 kDa 6 kDa
Insulin B Chain Unresolved Insulin 3.5 kDa
Insulin A Chain
2.5 kDa
SeeBlue
™
Pre-Stained
Standard
Tris-Glycine Gel (4–20%) Tricine Gel (10–20%)
Myosin 250 kDa 210 kDa
BSA 98 kDa 78 kDa
Glutamic Dehydrogenase 64 kDa 55 kDa
Alcohol Dehydrogenase 50 kDa 45 kDa
Carbonic Anhydrase 36 kDa 34 kDa
Myoglobin 30 kDa 23 kDa
Lysozyme 16 kDa 16 kDa
Aprotinin 6 kDa 7 kDa
Insulin 4 kDa 4 kDa
SeeBlue
™
Plus2 Pre-
Stained Standard
Tris-Glycine Gel (4–20%) Tricine Gel (10–20%)
Myosin 250 kDa 210 kDa
Phosphorylase B 148 kDa 105 kDa
BSA 98 kDa 78 kDa
Glutamic Dehydrogenase 64 kDa 55 kDa
Alcohol Dehydrogenase 50 kDa 45 kDa
Carbonic Anhydrase 36 kDa 34 kDa
Myoglobin 22 kDa 17 kDa
Lysozyme 16 kDa 16 kDa
Aprotinin 6 kDa 7 kDa
Insulin 4 kDa 4 kDa
Methods
Calibrating protein molecular weight
Novex
™
Pre-Cast gel electrophoresis guide User Guide
77
Troubleshooting
Review the information below to troubleshoot your experiments with Novex
™
Gels.
Observation Cause Solution
Run taking longer time Running buffer too dilute Make fresh running buffer as
described in this manual and avoid
adjusting the pH of the 1X running
buffer.
Low or no current during the run Incomplete circuit
•
Remove the tape from the
bottom of the cassette prior to
electrophoresis.
•
Make sure the buffer covers the
sample wells.
•
Check the wire connections on
the buffer core to make sure the
connections are intact.
Faint shadow or “ghost” band below
the expected protein band
Ghost bands are caused due to a
slight lifting of the gel from the
cassette resulting in trickling of some
sample beyond its normal migration
point. Gel lifting off the cassette is
caused due to:
•
Expired gels
•
Improper storage of gels
•
Avoid using expired gels. Use
fresh gels
•
Store the gels at the appropriate
temperature (see “Storage and
shelf life” on page 9).
Streaking of proteins
•
Sample overload
•
High salt concentration in the
sample
•
Sample precipitates
•
Contaminants such as
membranes or DNA complexes
in the sample
•
Load the appropriate amount of
protein as described on
“Recommended loading
volumes” on page 18.
•
Decrease the salt concentration
of your sample using dialysis or
gel ltration
•
Increase the concentration of
SDS in your sample if necessary,
to maintain the solubility of the
protein.
•
Centrifuge or clarify your sample
to remove particulate
contaminants
Methods
Troubleshooting
78
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Observation Cause Solution
Bands in the outer lane of the gel are
curving upwards
•
Concentrated buffer used
•
Expired gels used
•
High voltage used
•
The pre-made buffers are
supplied as concentrate. Dilute
the buffers as described in this
manual.
•
Avoid using gels after the
expiration date.
•
Electrophorese the gel using
conditions described on
“Electrophoresis conditions” on
page 47.
Bands in the outside lanes of the gel
“smiling”
Expired gels used causing the
acrylamide to break down in the gel
Avoid using gels after the expiration
date. Use fresh gels.
Bands are running as U shape rather
than a at band
Samples are loaded on the gel and
not electrophoresed immediately
resulting in sample diffusion
Load samples on to the gel
immediately before electrophoresis.
Bands appear to be “funneling” or
getting narrower as they progress
down the gel
Proteins are over-reduced causing
the proteins to be negatively charged
and repel each other.
Reduce the proteins using DTT or β-
mercaptoethanol as described on
“Reducing agent” on page 21.
Dumbbell shaped bands after
electrophoresis
Loading a large volume of sample
causing incomplete stacking of the
entire sample. This effect is
intensied for larger proteins
Load the appropriate volume of
sample per well as described on
“Recommended loading volumes” on
page 18. If your sample is too dilute,
concentrate the sample using salt
precipitation or ultraltration.
For TBE-Urea gels
High background and smeared
bands or abnormal band shapes
•
RNase contamination
•
Sample renatured
•
Sample overloaded
•
Urea not completely ushed
from the wells
•
Always wear gloves and use
sterile techniques to prevent
RNase contamination.
•
Heat the sample for 3 minutes at
70℃ and keep the sample in ice
to prevent renaturation. Proceed
to electrophoresis immediately
after loading.
•
Recommended DNA load is
0.16–0.33 µg/band.
•
Be sure to thoroughly ush urea
out of the wells prior to loading
the sample.
Methods
Troubleshooting
Novex
™
Pre-Cast gel electrophoresis guide User Guide
79
Appendix
Accessory products
Ordering information on a variety of electrophoresis reagents and apparatus available
from Thermo Fisher Scientic is provided below. For more information, visit our
website or call Technical Support.
Product Quantity Catalog no.
XCell
™
Mini-Cell
™
1 unit EI0001
XCell II Blot Module 1 unit EI9051
PowerEase
™
500 Power
Supply
1 unit EI8600
DryEase
™
Mini-Gel Drying
System
1 kit NI2387
StainEase
™
Staining Tray 2/pack NI2400
Gel-Dry
™
Drying Solution 500 mL LC4025
iBlot
™
Gel Transfer Device 1 unit IB1001
Novex
™
Semi-Dry Blotter 1 unit SD1000
Novex
™
Tris-Glycine SDS
Running Buffer (10X)
500 mL LC2675
NuPAGE
™
Sample Reducing
Agent (10X)
250 µL NP0004
NuPAGE
™
LDS Sample
Buffer (4X)
250 mL NP0008
Novex
™
Tris-Glycine
Transfer Buffer (25X)
500 mL LC3675
Novex
™
Tris-Glycine Native
Running Buffer (10X)
500 mL LC2672
Novex
™
Tris-Glycine Native
Sample Buffer (2X)
20 mL LC2673
Novex
™
Tris-Glycine SDS
Sample Buffer (2X)
20 mL LC2676
A
Electrophoresis
reagents
80
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Product Quantity Catalog no.
Novex
™
Tricine SDS
Running Buffer (10X)
500 mL LC1675
Novex
™
Tricine SDS Sample
Buffer (2X)
20 mL LC1676
Novex
™
Zymogram
Renaturing Buffer (10X)
500 mL LC2670
Novex
™
Zymogram
Developing Buffer (10X)
500 mL LC2671
Novex
™
TBE Running Buffer
(5X)
1 L LC6675
Novex
™
Hi-Density TBE
Sample Buffer (5X)
10 mL LC6678
Novex
™
TBE-Urea Sample
Buffer (2X)
10 mL LC6876
Novex
™
Prep TBE-Urea
Sample Buffer (2X)
20 mL LC6877
NuPAGE
™
Novex
™
4-12%
Bis-Tris ZOOM
™
Gel
1 gel NP0330BOX
Novex
™
4-20% Tris-Glycine
ZOOM
™
Gel
1 gel EC60261BOX
Novex
™
pH 3-7 IEF Buffer
Kit (includes LC5300,
LC5370, LC5371)
1 kit LC5377
Novex
™
pH 3-10 IEF Buffer
Kit (includes LC5300,
LC5310, LC5311)
1 kit LC5317
UltraPure
™
Agarose 100 g 15510-019
Nitrocellulose (0.45µm) 20 membrane/lter papers LC2000
Invitrolon
™
PVDF (0.45 µm) 20 membrane/lter papers LC2005
Nylon (0.45 µm) 20 membrane/lter papers LC2003
Appendix A Appendix
Accessory products
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
81
Ordering information for stains and protein molecular weight standards is provided
below. For more information, visit our website or contact Technical Support.
Product Application Quantity Catalog no.
SimplyBlue
™
Safe-
Stain
Fast, sensitive, safe
Coomassie
™
G-250
staining of proteins
in polyacrylamide
gels
1 L LC6060
SilverQuest
™
Silver
Staining Kit
Sensitive silver
staining of proteins
compatible with
mass spectrometry
analysis
1 Kit LC6070
Colloidal Blue
Staining Kit
Sensitive colloidal
Coomassie
™
G-250
staining of proteins
in polyacrylamide
gels
1 Kit LC6025
SilverXpress
™
Silver
Staining Kit
High-sensitivity, low
background protein
and nucleic acid
silver staining
1 Kit LC6100
Mark12
™
Unstained
Standard
For estimating the
apparent molecular
weight of proteins
1 mL LC5677
MagicMark
™
Western Standard
For protein
molecular weight
estimation on
western blots
250 µL LC5600
SeeBlue
™
Pre-
Stained Standard
For monitoring the
progress of your run
and evaluating
transfer efciency
500 µL LC5625
SeeBlue
™
Plus2 Pre-
Stained Standard
For visualizing
protein molecular
weight range and
evaluating transfer
efciency
500 µL LC5925
Novex
™
Sharp Pre-
stained Protein
Standard
For visualizing
protein molecular
weight range and
evaluating transfer
efciency
2 × 250 µL LC5800
Protein stains
and standards
Appendix A Appendix
Accessory products
A
82
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Product Application Quantity Catalog no.
BenchMark
™
Protein Ladder
For estimating the
apparent molecular
weight of proteins
2 × 250 µL 10747-012
IEF Marker 3-10 For determining the
pI of proteins
500 µL 39212-01
A large variety of nucleic acid markers are available from Thermo Fisher Scientic.
Ready-Load
™
format (pre-mixed with loading buffer) nucleic acid markers are also
available for your convenience. For more information, visit our website or contact
Technical Support.
Nucleic acid
markers
Appendix A Appendix
Accessory products
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
83
Recipes
The Tris-Glycine SDS Running Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
25 mM Tris Base
192 mM Glycine
0.1% SDS
pH 8.3
1.
To prepare 1,000 mL of 10X Tris-Glycine SDS Running Buffer, dissolve the
following reagents to 900 mL ultrapure water:
Tris Base 29 g
Glycine 144 g
SDS 10 g
2.
Mix well and adjust the volume to 1,000 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For electrophoresis, dilute this buffer to 1X with water (see “Preparing running
buffer” on page 23). The pH of the 1X solution is 8.3. Do not use acid or base to
adjust the pH.
The Tris-Glycine Native Running Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
25 mM Tris base
192 mM Glycine
pH 8.3
1.
To prepare 1,000 mL of 10X Tris-Glycine Native Running Buffer, dissolve the
following reagents to 900 mL ultrapure water:
Tris Base 29 g
Glycine 144 g
2.
Mix well and adjust the volume to 1,000 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For native electrophoresis, dilute this buffer to 1X with water (see “Preparing
running buffer” on page 23). The pH of the 1X solution is 8.3. Do not use acid or
base to adjust the pH.
Tris-Glycine
SDS running
buffer
Tris-Glycine
native running
buffer
Appendix A Appendix
Recipes
A
84
Novex
™
Pre-Cast gel electrophoresis guide User Guide
The Tris-Glycine SDS Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
63 mM Tris HCl
10% Glycerol
2% SDS
0.0025% Bromophenol Blue
pH 6.8
1.
To prepare 10 mL of 2X Tris-Glycine SDS Sample Buffer, mix the following
reagents :
0.5 M Tris-HCl, pH 6.8 2.5 mL
Glycerol 2 mL
10% (w/v) SDS 4 mL
0.1% (w/v) Bromophenol Blue 0.5 mL
2.
Adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
The Tris-Glycine Native Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
1X composition
100 mM Tris HCl
10% Glycerol
0.0025% Bromophenol Blue
pH 8.6
1.
To prepare 10 mL of 2X Tris-Glycine Native Sample Buffer, mix the following
reagents :
0.5 M Tris HCl, pH 8.6 4 mL
Glycerol 2 mL
0.1% (w/v) Bromophenol Blue 0.5 mL
2.
Adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
Tris-Glycine
SDS sample
buffer
Tris-Glycine
native sample
buffer
Appendix A Appendix
Recipes
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
85
The Tris-Glycine Transfer Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
12 mM Tris Base
96 mM Glycine
pH 8.3
1.
To prepare 500 mL of 25 × Tris-Glycine Transfer Buffer, dissolve the following
reagents in 400 mL ultrapure water:
Tris Base 18.2 g
Glycine 90 g
2.
Mix well and adjust the volume to 500 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For blotting, dilute this buffer as described on “Preparing transfer buffer” on
page 69. The pH of the 1X solution is 8.3. Do not use acid or base to adjust the
pH.
The Tricine SDS Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
450 mM Tris HCl
12% Glycerol
4% SDS
0.0025% Coomassie
™
Blue G
0.0025% Phenol Red
pH 8.45
1.
To prepare 10 mL of 2X Tricine SDS Sample Buffer, mix the following reagents:
3 M Tris HCl, pH 8.45 3 mL
Glycerol 2.4 mL
SDS 0.8 g
0.1% Coomassie
™
Blue G 0.5 mL
0.1% Phenol Red 0.5 mL
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
Tris-Glycine
transfer buffer
Tricine SDS
sample buffer
Appendix A Appendix
Recipes
A
86
Novex
™
Pre-Cast gel electrophoresis guide User Guide

The Tricine SDS Running Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
100 mM Tris base
100 mM Tricine
0.1% SDS
pH 8.3
1.
To prepare 1,000 mL of 10 × Tricine SDS Running Buffer, dissolve the following
reagents in 900 mL deionized water:
Tris Base 121 g
Tricine 179 g
SDS 10 g
2.
Mix well and adjust the volume to 1,000 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For electrophoresis, dilute this buffer to 1X with water (see “Preparing running
buffer” on page 26). The pH of the 1X solution is 8.3. Do not use acid or base to
adjust the pH.
The Zymogram Renaturing Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80). 25% (v/v) Triton
™
X-100
1.
To prepare 500 mL of 10X Zymogram Renaturing Buffer, add 125 mL Triton
™
X-100 to 300 mL ultra pure water.
2.
Mix well and adjust the volume to 500 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
The Zymogram Developing Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
50 mM Tris base
40 mM HCl
200 mM NaCl
5 mM CaCl
2
Tricine SDS
running buffer
10X zymogram
renaturing
buffer
Zymogram
developing
buffer
Appendix A Appendix
Recipes
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
87
0.02% (w/v) Brij
™
35
1.
To prepare 500 mL of 10X Zymogram Developing Buffer, dissolve the following
reagents in 400 mL deionized water:
Tris Base 30.2 g
6N HCl 33 mL
NaCl 58.5 g
CaCl
2
.2H
2
O 3.7 g
Brij
™
35 1.0 g
2.
Mix well and adjust the volume to 500 mL with ultrapure water.
3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For developing the zymogram gel, dilute this buffer to 1X with water (see
“Preparing developing buffer” on page 29).
The IEF Sample Buffer pH 3–7 is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
40 mM Lysine (free base)
15% Glycerol
1.
To prepare 10 mL of 2X IEF Sample Buffer pH 3–7, mix the following reagents:
10X IEF Cathode Buffer, pH 3–7 2 mL
Glycerol 3 mL
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
The IEF Sample Buffer pH 3–10 is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
20 mM Lysine (free base)
20 mM Arginine (free base)
15% Glycerol
1.
To prepare 10 mL of 2X IEF Sample Buffer pH 3–10, mix the following reagents:
10X IEF Cathode Buffer, pH 3–10
2 mL
Glycerol 3 mL
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
IEF sample
buffer pH 3–7
IEF sample
buffer, pH 3–10
Appendix A Appendix
Recipes
A
88
Novex
™
Pre-Cast gel electrophoresis guide User Guide

The IEF Cathode Buffer pH 3–7 is available from Thermo Fisher Scientic (see
“Accessory products” on page 80). 40 mM Lysine (free base)
1.
To prepare 100 mL of 10X IEF Cathode Buffer pH 3–7, dissolve 5.8 g of Lysine
(free base) in 100 mL of ultrapure water.
2.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
The IEF Cathode Buffer pH 3–10 is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
20 mM Lysine (free base)
20 mM Arginine (free base) You can use D, L, or D/L form of arginine
pH 10.1
1.
To prepare 100 mL of 10X IEF Cathode Buffer pH 3–10, dissolve 2.9 g of Lysine
(free base) and 3.5 g of Arginine (free base) in 100 mL of ultrapure water.
2.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
The IEF Anode Buffer is available from Thermo Fisher Scientic (see “Accessory
products” on page 80). 7 mM Phosphoric acid
1.
To prepare 100 mL of 50X IEF Anode Buffer, mix 2.4 mL of 85% phosphoric acid
with 97.6 mL of ultrapure water.
2.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
The TBE Running Buffer is available from Thermo Fisher Scientic (see “Accessory
products” on page 80).
89 mM Tris base
89 mM Boric acid
2 mM EDTA (free acid)
pH 8.3
1.
To prepare 1,000 mL of 5X TBE Running Buffer, dissolve the following reagents
in 900 mL deionized water:
Tris Base 54 g
Boric acid 27.5 g
EDTA (free acid) 2.9 g
2.
Mix well and adjust the volume to 1,000 mL with ultrapure water.
IEF cathode
buffer, pH 3–7
IEF cathode
buffer, pH 3–10
IEF anode
buffer
TBE running
buffer
Appendix A Appendix
Recipes
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
89

3.
Store at room temperature. The buffer is stable for 6 months when stored at
room temperature.
4.
For electrophoresis, dilute this buffer to 1X with water as described on
“Preparing running buffer” on page 39. The pH of the 1X solution is 8.3. Do not
use acid or base to adjust the pH.
The Hi-Density TBE Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
18 mM Tris base
18 mM Boric acid
0.4 mM EDTA (free acid)
3% Ficoll
™
Type 400
0.02% Bromophenol Blue
0.02% Xylene Cyanol
1.
To prepare 10 mL of 5X Hi-Density TBE Sample Buffer, dissolve the following
reagents in 9 mL deionized water:
5X TBE Running Buffer (see ) 2 mL
Ficoll
™
Type 400 1.5 g
1% Bromophenol Blue 1 mL
1% Xylene Cyanol 1 mL
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
The TBE-Urea Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
45 mM Tris base
45 mM Boric acid
1 mM EDTA (free acid)
6% Ficoll
™
Type 400
3.5 M Urea
0.005% Bromophenol Blue
Hi-Density TBE
sample buffer
TBE-Urea
sample buffer
Appendix A Appendix
Recipes
A
90
Novex
™
Pre-Cast gel electrophoresis guide User Guide
0.025% Xylene Cyanol
1.
To prepare 10 mL of 2X TBE-Urea Sample Buffer, dissolve the following reagents
in 9 mL deionized water:
5X TBE Running Buffer 2 mL
Ficoll
™
Type 400 1.2 g
1% Bromophenol Blue 1 mL
1% Xylene Cyanol 0.5 mL
Urea 4.2 g
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 3 months when stored at +4℃.
The Prep TBE–Urea Sample Buffer is available from Thermo Fisher Scientic (see
“Accessory products” on page 80).
45 mM Tris base
45 mM Boric acid
1 mM EDTA (free acid)
6% Ficoll
™
Type 400
3.5 M Urea
1.
To prepare 10 mL of 2X Prep TBE–Urea Sample Buffer, dissolve the following
reagents in 9 mL deionized water:
5X TBE Running Buffer (see “TBE
running buffer” on page 89)
2 mL
Ficoll
™
Type 400 1.2 g
Urea 4.2 g
2.
Mix well and adjust the volume to 10 mL with ultrapure water.
3.
Store at +4℃. The buffer is stable for 6 months when stored at +4℃.
Prep TBE–Urea
sample buffer
Appendix A Appendix
Recipes
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
91
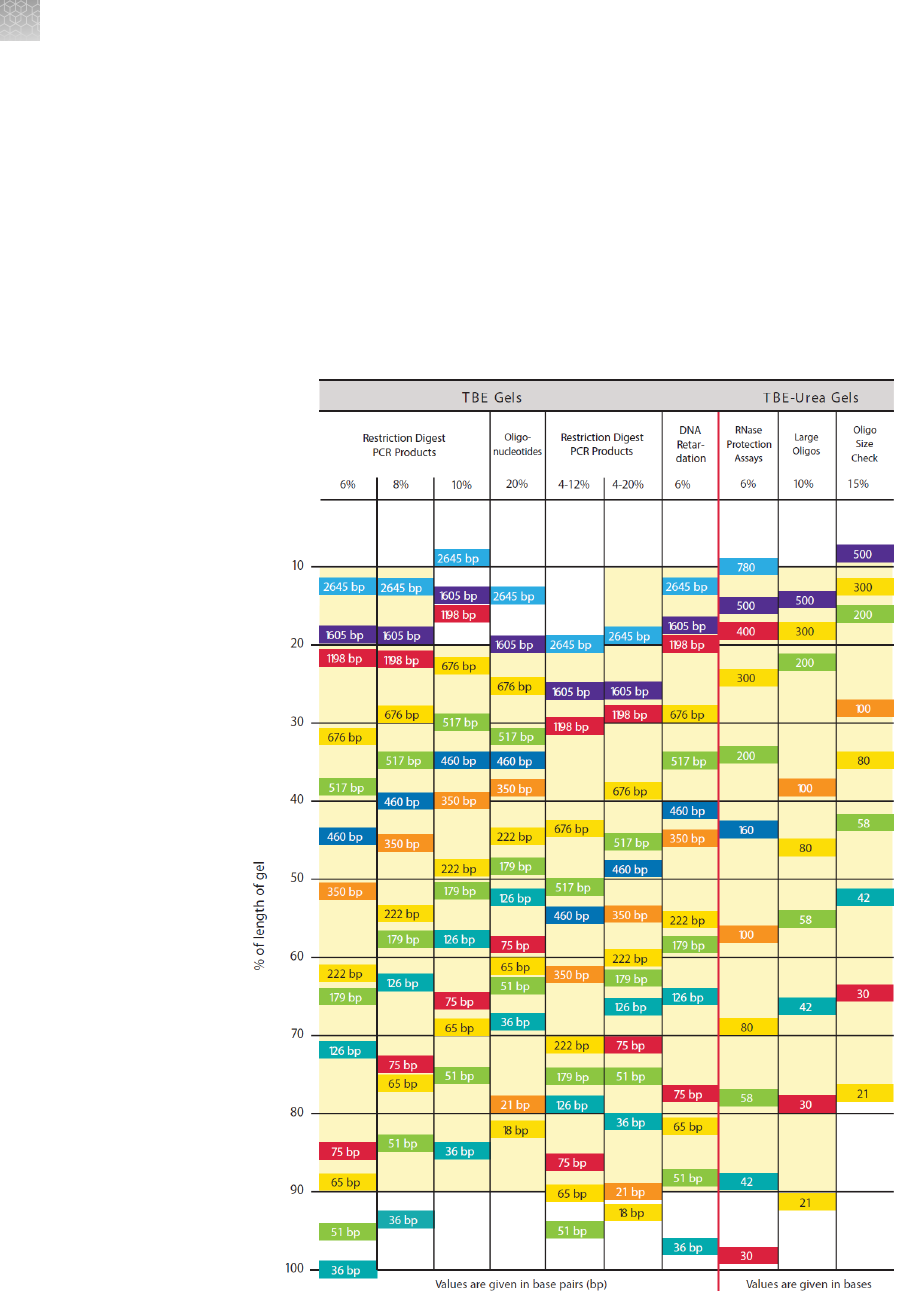
Gel migration charts
The migration patterns of protein standards on Novex
™
Tris-Glycine Gels are shown
on the table below. Use the table to select the proper gel for separating proteins
based on size. Optimal resolution is achieved when protein bands migrate within the
shaded regions.
Note: Bands correspond to the migration of Mark12
™
Unstained Standard under
denaturing conditions.
Novex
™
Tris-
Glycine gel
migration chart
Appendix A Appendix
Gel migration charts
A
92
Novex
™
Pre-Cast gel electrophoresis guide User Guide
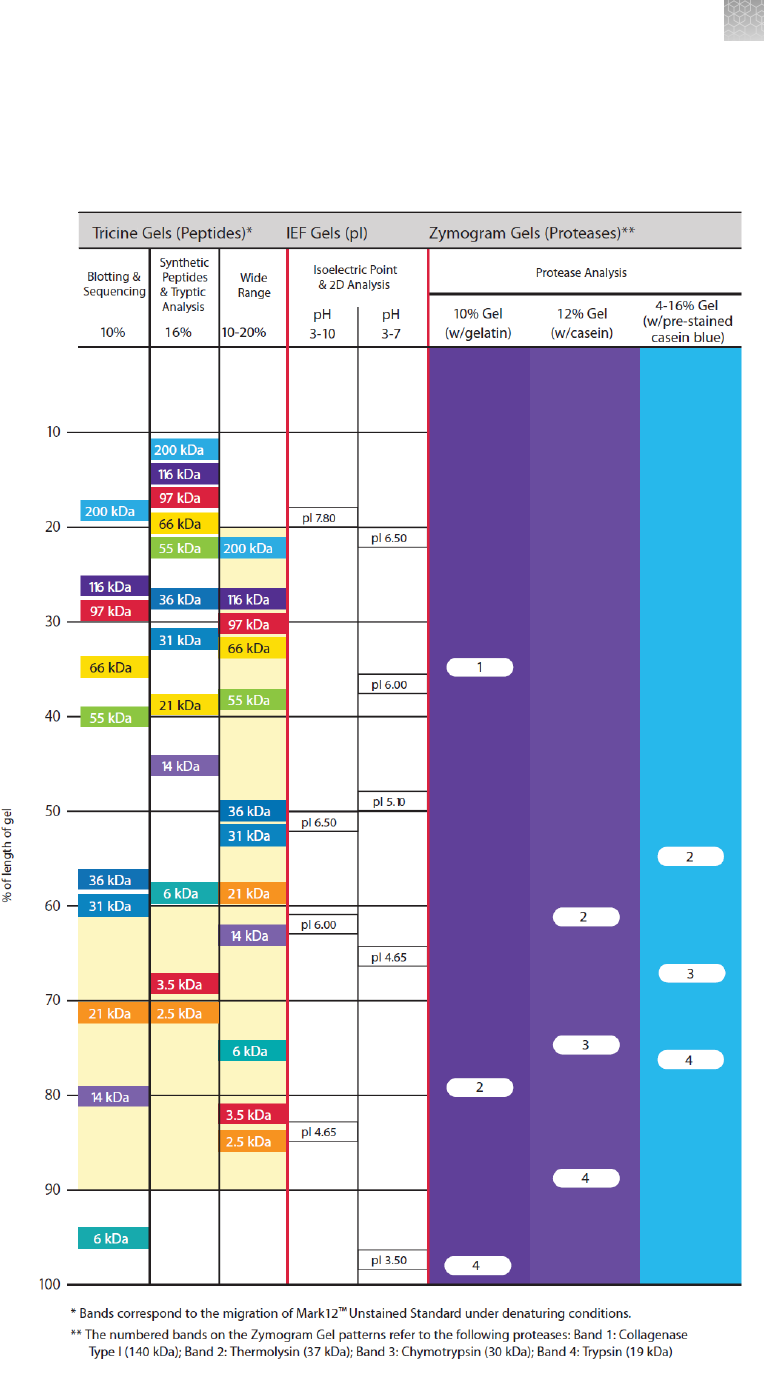
The migration patterns of protein markers on Novex
™
Tricine, IEF, and Zymogram
Gels are shown on the table below. Optimal resolution is achieved when protein
bands migrate within the shaded regions.
Novex
™
tricine,
IEF, and
zymogram gel
migration chart
Appendix A Appendix
Gel migration charts
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
93

The migration patterns of DNA fragments on Novex
™
TBE and TBE-Urea Gels are
shown on the table below. Optimal resolution is achieved when nucleic acid bands
migrate within the shaded regions.
Novex
™
TBE
and TBE-Urea
gel migration
chart
Appendix A Appendix
Gel migration charts
A
94
Novex
™
Pre-Cast gel electrophoresis guide User Guide
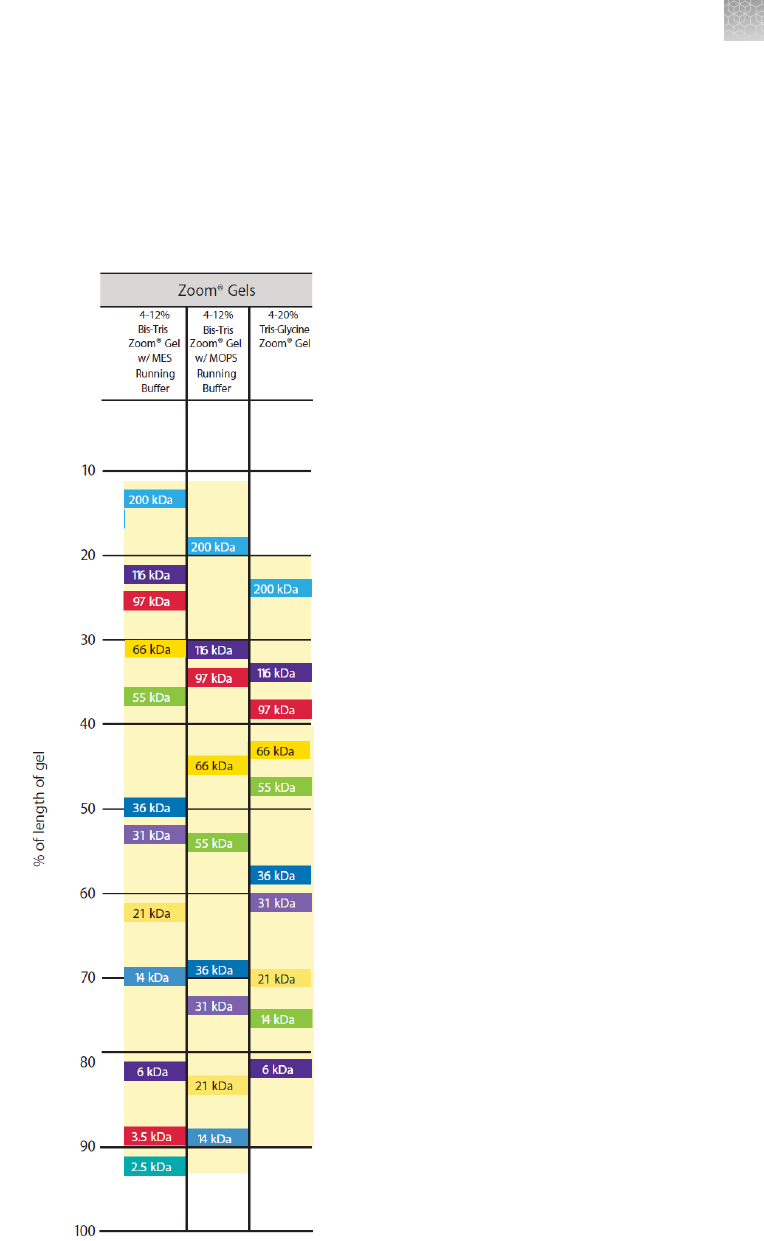
The migration patterns of protein standards on ZOOM
™
Gels are shown on the table
below. Optimal resolution is achieved when protein bands migrate within the shaded
regions.
Note: On ZOOM
™
Gels, migration of bands correspond to the migration of Mark12
™
Unstained Standard (Cat. no. LC5677) under denaturing conditions.
ZOOM
™
gel
migration chart
Appendix A Appendix
Gel migration charts
A
Novex
™
Pre-Cast gel electrophoresis guide User Guide
95

References
Kubo, K. (1995). Effect of Incubation of Solutions of Proteins Containing Dodecyl
Sulfate on the Cleavage of Peptide Bonds by Boiling. Anal. Biochem. 225, 351-353.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the
head of bacteriophage T4. Nature 227, 680-685.
Ornstein, L. (1964). Disc Electrophoresis, 1, Background and Theory. Ann New York
Acad. Sci 121, 321-349.
Revzin, A. (1989). Gel Electrophoresis Assays for DNA-Protein Interactions.
BioTechniques 4, 346-355.
Schaegger, H., and von Jagow, G. (1987). Tricine-Sodium dodecyl sulfate-
Polyacrylamide Gel Electrophoresis for the Separation of Proteins in the Range from
1 to 100 kDa. Anal. Biochem. 166, 368-379.
Appendix A Appendix
References
A
96
Novex
™
Pre-Cast gel electrophoresis guide User Guide

Safety
WARNING! GENERAL SAFETY. Using this product in a manner not specied in the
user documentation may result in personal injury or damage to the instrument or
device. Ensure that anyone using this product has received instructions in general
safety practices for laboratories and the safety information provided in this
document.
·
Before using an instrument or device, read and understand the safety information
provided in the user documentation provided by the manufacturer of the
instrument or device.
·
Before handling chemicals, read and understand all applicable Safety Data Sheets
(SDSs) and use appropriate personal protective equipment (gloves, gowns, eye
protection, and so on). To obtain SDSs, see the “Documentation and Support”
section in this document.
B
Novex
™
Pre-Cast gel electrophoresis guide User Guide
97

Chemical safety
WARNING! GENERAL CHEMICAL HANDLING. To minimize hazards, ensure
laboratory personnel read and practice the general safety guidelines for chemical
usage, storage, and waste provided below. Consult the relevant SDS for specic
precautions and instructions:
·
Read and understand the Safety Data Sheets (SDSs) provided by the chemical
manufacturer before you store, handle, or work with any chemicals or hazardous
materials. To obtain SDSs, see the "Documentation and Support" section in this
document.
·
Minimize contact with chemicals. Wear appropriate personal protective equipment
when handling chemicals (for example, safety glasses, gloves, or protective
clothing).
·
Minimize the inhalation of chemicals. Do not leave chemical containers open. Use
only with sufcient ventilation (for example, fume hood).
·
Check regularly for chemical leaks or spills. If a leak or spill occurs, follow the
manufacturer cleanup procedures as recommended in the SDS.
·
Handle chemical wastes in a fume hood.
·
Ensure use of primary and secondary waste containers. (A primary waste container
holds the immediate waste. A secondary container contains spills or leaks from the
primary container. Both containers must be compatible with the waste material
and meet federal, state, and local requirements for container storage.)
·
After emptying a waste container, seal it with the cap provided.
·
Characterize (by analysis if needed) the waste generated by the particular
applications, reagents, and substrates used in your laboratory.
·
Ensure that the waste is stored, transferred, transported, and disposed of
according to all local, state/provincial, and/or national regulations.
·
IMPORTANT! Radioactive or biohazardous materials may require special handling,
and disposal limitations may apply.
WARNING! HAZARDOUS WASTE (from instruments). Waste produced by the
instrument is potentially hazardous. Follow the guidelines noted in the preceding
General Chemical Handling warning.
WARNING! 4L Reagent and Waste Bottle Safety. Four-liter reagent and waste
bottles can crack and leak. Each 4-liter bottle should be secured in a low-density
polyethylene safety container with the cover fastened and the handles locked in the
upright position.
Appendix B Safety
Chemical safety
B
98
Novex
™
Pre-Cast gel electrophoresis guide User Guide
Biological hazard safety
WARNING! Potential Biohazard. Depending on the samples used on this
instrument, the surface may be considered a biohazard. Use appropriate
decontamination methods when working with biohazards.
WARNING! BIOHAZARD. Biological samples such as tissues, body uids,
infectious agents, and blood of humans and other animals have the potential to
transmit infectious diseases. Conduct all work in properly equipped facilities with the
appropriate safety equipment (for example, physical containment devices). Safety
equipment can also include items for personal protection, such as gloves, coats,
gowns, shoe covers, boots, respirators, face shields, safety glasses, or goggles.
Individuals should be trained according to applicable regulatory and company/
institution requirements before working with potentially biohazardous materials.
Follow all applicable local, state/provincial, and/or national regulations. The following
references provide general guidelines when handling biological samples in laboratory
environment.
·
U.S. Department of Health and Human Services, Biosafety in Microbiological and
Biomedical Laboratories (BMBL), 5th Edition, HHS Publication No. (CDC) 21-1112,
Revised December 2009; found at:
https://www.cdc.gov/labs/pdf/
CDC-BiosafetymicrobiologicalBiomedicalLaboratories-2009-P.pdf
·
World Health Organization, Laboratory Biosafety Manual, 3rd Edition,
WHO/CDS/CSR/LYO/2004.11; found at:
www.who.int/csr/resources/publications/biosafety/Biosafety7.pdf
Appendix B Safety
Biological hazard safety
B
Novex
™
Pre-Cast gel electrophoresis guide User Guide
99
Documentation and support
Customer and technical support
Visit
thermofisher.com/support
for the latest service and support information.
•
Worldwide contact telephone numbers
•
Product support information
–
Product FAQs
–
Software, patches, and updates
–
Training for many applications and instruments
•
Order and web support
•
Product documentation
–
User guides, manuals, and protocols
–
Certicates of Analysis
–
Safety Data Sheets (SDSs; also known as MSDSs)
Note: For SDSs for reagents and chemicals from other manufacturers,
contact the manufacturer.
Limited product warranty
Life Technologies Corporation and/or its afliate(s) warrant their products as set forth
in the Life Technologies' General Terms and Conditions of Sale at
www.thermofisher.com/us/en/home/global/terms-and-conditions.html. If you
have any questions, please contact Life Technologies at www.thermofisher.com/
support.
100
Novex
™
Pre-Cast gel electrophoresis guide User Guide

