
Indexed Sequencing
Overview Guide
Document # 15057455 v08
November 2020
ILLUMINA PROPRIETARY
For Research Use Only. Not for use in diagnostic procedures.
This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"), and are intended solely
for the contractual use of its customer in connection with the use of the product(s) described herein and for no other
purpose. This document and its contents shall not be used or distributed for any other purpose and/or otherwise
communicated, disclosed, or reproduced in any way whatsoever without the prior written consent of Illumina. Illumina
does not convey any license under its patent, trademark, copyright, or common-law rights nor similar rights of any
third parties by this document.
The instructions in this document must be strictly and explicitly followed by qualified and properly trained personnel
in order to ensure the proper and safe use of the product(s) described herein. All of the contents of this document
must be fully read and understood prior to using such product(s).
FAILURE TO COMPLETELY READ AND EXPLICITLY FOLLOW ALL OF THE INSTRUCTIONS CONTAINED HEREIN MAY
RESULT IN DAMAGE TO THE PRODUCT(S), INJURY TO PERSONS, INCLUDING TO USERS OR OTHERS, AND DAMAGE
TO OTHER PROPERTY, AND WILL VOID ANY WARRANTY APPLICABLE TO THE PRODUCT(S).
ILLUMINA DOES NOT ASSUME ANY LIABILITY ARISING OUT OF THE IMPROPER USE OF THEPRODUCT(S)
DESCRIBED HEREIN (INCLUDING PARTS THEREOF OR SOFTWARE).
© 2020 Illumina, Inc. All rights reserved.
All trademarks are the property of Illumina, Inc. or their respective owners. For specific trademark information, see
www.illumina.com/company/legal.html.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
ii
Indexed Sequencing Overview Guide

Revision History
Document Date Description of Change
Document # 15057455
v08
November
2020
Updates made to support the MiniSeq Standard and Rapid
reagent kits.
Document # 15057455
v07
July 2020 Updates made to support the NovaSeq 6000 v1.5 reagent
kit introduction.
Document # 15057455
v06
March
2020
Modified NextSeq Systems reference to account for all
versions.
Added Instrument Run Setup to introduction.
Document # 15057455
v05
March
2019
Renamed the two dual-indexed workflows on a paired-end
flow cell:
• Renamed workflow A to the forward strand workflow.
• Renamed workflow B to the reverse complement
workflow.
Updated cycles per Index Read for all dual-index
workflows (except forward strand) to eight or 10.
Updated the descriptions of single- and dual-indexed
libraries.
Corrected the Index 1primer for the HiSeq 3000/4000
SR Cluster Kit, HiSeq SR Cluster Kit v4, and HiSeq
PE Cluster Kit v4 to HP12.
Document # 15057455
v04
February
2018
Added the iSeq 100 and HiSeq X flow cells to workflow B
for dual-indexing on a paired-end flow cell.
Added the IDT for Illumina TruSeq UD Indexes
combinations for dual-indexed libraries.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
iii
Indexed Sequencing Overview Guide

Document Date Description of Change
Document # 15057455
v03
February
2017
Updated for the NovaSeq Series:
• Added the NovaSeq 5000/6000 Flow Cell to workflow A
for dual-indexing on a paired-end flow cell.
• For workflow A, increased the number of cycles in an
Index Read to a maximum of 20.
Updated how many uniquely tagged libraries can be
generated:
• Up to 48 single-indexed libraries.
• Up to 384 dual-indexed libraries.
Clarified that this guide is applicable to all Illumina
sequencing systems.
Document # 15057455
v02
March
2016
Added the MiniSeq system, which follows the single-index
workflow and Workflow B for dual-indexing on a paired-
end flow cell.
Renamed this guide to Indexed Sequencing Overview
Guide to emphasize indexing over systems.
Organized dual-indexing workflows on paired-end flow
cells as Workflow A and Workflow B.
Organized dual-indexing workflows on single-read flow
cells by sequencing system.
Document # 15057455
v01
August
2015
Added the dual-indexed workflow for a HiSeq 3000/4000
SR flow cell.
Added sequencing primers available in the HiSeq
3000/4000 SR Cluster Kit.
Part # 15057455 Rev.
B
February
2015
Added the HiSeq 3000/4000 flow cell to the dual-indexed
workflow that performs the Index 2 Read after Read 2
resynthesis. This workflow is performed on NextSeq,
HiSeq 4000, and HiSeq 3000.
Added sequencing primers available in the HiSeq
3000/4000 PECluster Kit.
Part # 15057455 Rev.
A
July 2014 Initial release.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
iv
Indexed Sequencing Overview Guide

Table of Contents
Revision History iii
Introduction 1
Single and Dual Indexing 1
Single-Indexed Sequencing Overview 2
Dual-Indexed Sequencing Overview 2
Dual-Indexing Workflows 3
Dual-Indexed Workflow on a Paired-End Flow Cell 3
Forward Strand Workflow 4
Reverse Complement Workflow 5
Dual-Indexed Workflow on a Single-Read Flow Cell 6
HiSeq 4000 and HiSeq 3000 Systems 6
HiSeq 2500 and HiSeq 2000 Systems 7
Sequencing Primers for HiSeq Systems 7
Sequencing Primers in Cluster Kits 8
Additional Primers for the TruSeq Cluster Kit v3 8
Technical Assistance 9
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
v
Indexed Sequencing Overview Guide
Introduction
This guide provides an overview of indexed sequencing for Illumina sequencing systems. Indexed
sequencing is a method that allows multiple libraries to be pooled and sequenced together.
Indexing libraries requires the addition of a unique identifier, or index sequence, to DNA samples during
library preparation. BaseSpace Sequence Hub, Local Run Manager, Instrument Run Setup, or
bcl2fastq2 Conversion Software process these tags to identify each uniquely tagged library for
downstream analysis.
Single and Dual Indexing
The number of index sequences added to samples differs for single-indexed and dual-indexed
sequencing.
• Single-indexed libraries—Adds Index 1(i7) sequences to generate uniquely tagged libraries.
• Dual-indexed libraries—Adds Index 1(i7) and Index 2 (i5) sequences to generate uniquely tagged
libraries.
– Unique dual (UD) indexes have distinct, unrelated index adapters for both index reads. Index
adapter sequences are eight or 10 bases long.
– Combinatorial dual (CD) indexes have eight unique dual pairs of index adapters, so most
libraries share sequences on the i7 or i5 end. Index adapter sequences are eight bases long.
During indexed sequencing, the index is sequenced in a separate read called the Index Read, where a
new sequencing primer is annealed. When libraries are dual-indexed, the sequencing run includes two
additional reads, called the Index 1Read and Index 2 Read.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
1
Indexed Sequencing Overview Guide

Single-Indexed Sequencing Overview
The single-indexed sequencing workflow applies to all Illumina sequencing platforms, where an Index
Read follows Read 1.
Figure 1 Single-Indexed Sequencing
1. Read 1—Read 1follows the standard Read 1sequencing protocol using SBS reagents. The Read 1
sequencing primer is annealed to the template strand during the cluster generation step.
2. Index Read preparation—The Read 1product is removed and the Index 1(i7) sequencing primer is
annealed to the same template strand, producing the Index 1(i7) Read.
3. Index 1 (i7) Read—Following Index Read preparation, the Index 1(i7) Read is performed. The read
length depends on the system and run parameters.
4. Read 2 resynthesis—The Index Read product is removed and the original template strand is used
to regenerate the complementary strand. Then, the original template strand is removed to allow
hybridization of the Read 2 sequencing primer.
5. Read 2—Read 2 follows the standard paired-end sequencing protocol using SBS reagents.
Dual-Indexed Sequencing Overview
Dual-indexed sequencing includes two index reads after Read 1: the Index 1Read and the Index 2
Read.
Sequencing kits for HiSeq™systems are available with a single-read or paired-end flow cell. For all
other systems, sequencing kits include a paired-end flow cell.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
2
Indexed Sequencing Overview Guide

Dual-Indexing Workflows
The control software performs Read 1, any index reads, and then Read 2 based on the parameters
provided for the run in the sample sheet or during run setup.
For all indexing workflows, the Index 1Read directly follows Read 1. However, for dual-indexing on a
paired-end flow cell, the rest of the workflow differs:
• Forward strand—The Index 2 Read occurs before Read 2 resynthesis, so the Index 2 (i5) adapter is
sequenced on the forward strand.
• Reverse complement—The Index 2 Read occurs after Read 2 resynthesis, which creates the
reverse complement of the Index 2 (i5) index adapter sequence.
Forward Strand Reverse Complement
Read 1
Index Read preparation
Index 1Read
Index 2 Read
Read 2 resynthesis
Read 2
Read 1
Index Read preparation
Index 1Read
Read 2 resynthesis
Index 2 Read
Read 2 preparation
Read 2
Table 1 Dual-Index Paired-End Sequencing Workflows
Dual-Indexed Workflow on a Paired-End Flow
Cell
Dual-index sequencing on a paired-end flow cell follows one of two workflows, depending on the
system and software:
• The forward strand workflow is performed on the NovaSeq 6000 with v1.0 reagent kits, MiniSeq
with Rapid Reagent kits, MiSeq, HiSeq 2500, and HiSeq 2000.
• The reverse complement workflow is performed on the iSeq 100, MiniSeq with Standard reagent
kits, NextSeq Systems, NovaSeq 6000 with v1.5 reagent kits, HiSeq X, HiSeq 4000, and
HiSeq 3000.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
3
Indexed Sequencing Overview Guide

Forward Strand Workflow
The chemistry applied to the Index 2 Read during a paired-end, dual-indexed run on the NovaSeq 6000
with v1.0 reagent kits, MiniSeq with rapid reagent kits, MiSeq, HiSeq 2500, or HiSeq 2000 System is
specific to the paired-end flow cell. Reading the i5 index requires seven additional chemistry-only
cycles. This step uses the resynthesis mix, a paired-end reagent, during the Index 2 Read process.
NOTE: While MiniSeq Rapid reagents allow for dual indexing, Read 2 cannot be performed
with the MiniSeq Rapid reagent kit.
Figure 2 Dual-Indexed Sequencing on a Paired-End Flow Cell (Forward Strand)
1. Read 1—Read 1follows the standard Read 1sequencing protocol using SBS reagents. The Read 1
sequencing primer is annealed to the template strand during the cluster generation step.
2. Index Read preparation—The Read 1product is removed and the Index 1(i7) sequencing primer is
annealed to the same template strand.
3. Index 1 (i7) Read—Following Index Read preparation, the Index 1(i7) Read performs up to 20
cycles of sequencing.
The maximum number of cycles in each Index Read depends on the system and run
parameters.
4. Index 2 (i5) Read—The Index 1(i7) Read product is removed and the template anneals to the
grafted P5 primer on the surface of the flow cell. The run proceeds through an additional seven
chemistry-only cycles (no imaging occurs), followed by up to 20 cycles of sequencing.
5. Read 2 resynthesis—The Index Read product is removed and the original template strand is used
to regenerate the complementary strand. The original template strand is then removed to allow
hybridization of the Read 2 sequencing primer.
6. Read 2—Read 2 follows the standard paired-end sequencing protocol using SBS reagents.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
4
Indexed Sequencing Overview Guide

Reverse Complement Workflow
A dual-indexed run on the iSeq 100, MiniSeq with standard reagent kits, NextSeq Systems, NovaSeq
6000 with v1.5 reagent kits, HiSeq X, HiSeq 4000, or HiSeq 3000 System performs the Index 2 Read
after Read 2 resynthesis. This workflow requires a reverse complement of the Index 2 (i5) primer
sequence compared to the primer sequence used on other Illumina platforms.
The Index 2 sequencing primer is part of the dual-indexing primer mix for the iSeq 100, MiniSeq with
standard reagent kits, NextSeq Systems, and NovaSeq 6000 with v1.5 reagent kits. For the HiSeq X,
HiSeq 4000, and HiSeq 3000 Systems, the Index 2 sequencing primer is part of HP14. HP14 is an
indexing primer mix that contains primers for both index reads.
Figure 3 Dual-Indexed Sequencing on a Paired-End Flow Cell (Reverse Complement)
1. Read 1—Read 1follows the standard Read 1sequencing protocol using SBS reagents. The Read 1
sequencing primer is annealed to the template strand during the cluster generation step.
2. Index Read preparation—The Read 1product is removed and the Index 1(i7) sequencing primer is
annealed to the same template strand.
3. Index 1 (i7) Read—Following Index Read preparation, the Index 1(i7) Read performs eight or 10
cycles of sequencing.
4. Read 2 resynthesis—The Index 1Read product is removed and the original template strand is used
to regenerate the complementary strand. Then the original template strand is removed to allow
hybridization of the Index 2 (i5) sequencing primer.
5. Index 2 (i5) Read—Following Read 2 resynthesis, the Index 2 (i5) Read performs eight or 10 cycles
of sequencing.
This workflow does not require seven additional chemistry-only cycles.
6. Read 2 preparation—The Index 2 Read product is removed and the Read 2 sequencing primer is
annealed to the same template strand.
7. Read 2—Read 2 follows the standard paired-end sequencing protocol using SBS reagents.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
5
Indexed Sequencing Overview Guide

Dual-Indexed Workflow on a Single-Read
Flow Cell
Single-read sequencing is possible on all HiSeq systems. Dual-index sequencing on a single-read flow
cell follows one of two workflows, depending on the system.
HiSeq 4000 and HiSeq 3000 Systems
The chemistry applied to the Index 2 Read during a single-read dual-indexed run on the HiSeq 4000 or
HiSeq 3000 System is specific to the single-read flow cell. Reading the i5 index requires seven
additional chemistry-only cycles. This step uses the resynthesis mix during the Index 2 Read.
Figure 4 Dual-Indexed Sequencing on a Single-Read Flow Cell (HiSeq 4000 or HiSeq 3000)
1. Read 1—Read 1follows the standard Read 1sequencing protocol using SBS reagents. The Read 1
sequencing primer is annealed to the template strand during cluster generation.
2. Index Read preparation—The Read 1product is removed and the Index 1(i7) sequencing primer is
annealed to the same template strand.
3. Index 1 (i7) Read—Following Index Read preparation, the Index 1(i7) Read performs eight or 10
cycles of sequencing.
4. Index 2 (i5) Read—The Index 1(i7) Read product is removed and the template anneals to the
grafted P5 oligo on the surface of the flow cell. The run proceeds through seven chemistry-only
cycles (no imaging occurs), followed by eight or 10 cycles of sequencing.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
6
Indexed Sequencing Overview Guide
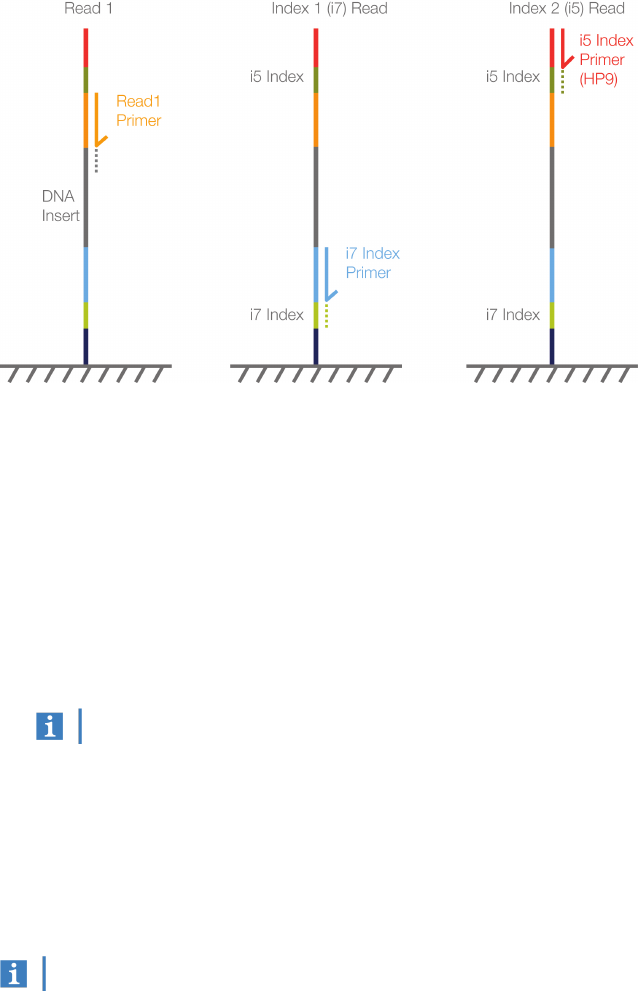
HiSeq 2500 and HiSeq 2000 Systems
The chemistry applied to the Index 2 Read during a single-read, dual-indexed run on the HiSeq 2500 or
HiSeq 2000 System is specific to the single-read flow cell. Performing the Index 2 Read on a HiSeq
single-read flow cell requires HP9, an Index 2 sequencing primer.
Figure 5 Dual-Indexed Sequencing on a Single-Read Flow Cell (HiSeq 2500 or HiSeq 2000)
1. Read 1—Read 1follows the standard Read 1sequencing protocol using SBS reagents. The Read 1
sequencing primer is annealed to the template strand during the cluster generation step.
2. Index Read preparation—The Read 1product is removed and the Index 1(i7) sequencing primer is
annealed to the same template strand.
3. Index 1 (i7) Read—Following Index Read preparation, the Index 1(i7) Read performs eight or 10
cycles of sequencing.
4. Index 2 (i5) Read—The Index 1(i7) Read product is removed and the Index 2 (i5) sequencing
primer is annealed to the same template strand. The run proceeds through eight or 10 cycles of
sequencing.
This workflow does not require seven additional chemistry-only cycles.
Sequencing Primers for HiSeq Systems
Indexing workflow differences require system-specific chemistry and sequencing primers. The
following tables list available HiSeq reagent kits and the associated sequencing primers, which are
used with each step of an indexed run.
Sequencing primers for all other systems are provided in the prefilled reagent cartridge.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
7
Indexed Sequencing Overview Guide

Sequencing Primers in Cluster Kits
Run Type Read 1 Index 1(i7) Index 2 (i5) Read 2
HiSeq 3000/4000 PECluster
Kit
HP10 HP14 HP14 HP11
HiSeq 3000/4000 SR Cluster
Kit
HP10 HP12 --¹ --
HiSeq PE Cluster Kit v4 HP10 HP12 --¹ HP11
HiSeq SR Cluster Kit v4 HP10 HP12 HP9 --
TruSeq PECluster Kit v3 HP6 HP8 --¹ HP7
TruSeq SRCluster Kit v3 HP6 HP8 --² --
¹ The Index 2 Read uses resynthesis mix.
² The TruSeq Dual Index Sequencing Primer Box for single reads is required for dual- indexed sequencing on a
single- read flow cell, regardless of library type.
Additional Primers for the TruSeq Cluster Kit v3
Using the TruSeq Cluster Kit v3 to sequence any Nextera libraries except Nextera Mate Pair libraries
requires the TruSeq Dual Index Sequencing Primer Box. Sequencing primers in TruSeq v3 kits are not
compatible with most Nextera libraries, while sequencing primers provided in the TruSeq Dual Index
Sequencing Primer Box are compatible with all library types. To confirm primer compatibility, see the
documentation for the library prep kit.
Dual-indexed sequencing on a single-read flow cell requires the single-read kit, regardless of the
libraries being sequenced.
Run Type Read 1 Index 1(i7) Index 2 (i5) Read 2
TruSeq PEDual Index
Sequencing Primer Box
(For use with paired-end flow
cells)
HP10 HP12 --¹ HP11
TruSeq SRDual Index
Sequencing Primer Box
(For use with single-read flow
cells)
HP10 HP12 HP9 --
¹ The resynthesis mix, a paired-end reagent provided in the TruSeq PE Cluster Kit v3, is used to perform the Index 2
Read.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
8
Indexed Sequencing Overview Guide

Technical Assistance
For technical assistance, contact Illumina Technical Support.
Website: www.illumina.com
Email: techsupport@illumina.com
Illumina Technical Support Telephone Numbers
Region Toll Free International
Australia +611800 775 688
Austria +43 800 006249 +43 19286540
Belgium +32 800 77 160 +32 3 400 29 73
Canada +1800 809 4566
China +86 400 066 5835
Denmark +45 80 82 0183 +45 89 87 1156
Finland +358 800 918 363 +358 9 7479 0110
France +33 8 05 10 2193 +33 170 77 04 46
Germany +49 800 1014940 +49 89 3803 5677
Hong Kong, China +852 800 960 230
India +918006500375
Indonesia 0078036510048
Ireland +353 1800 936608 +353 1695 0506
Italy +39 800 985513 +39 236003759
Japan +810800 1115011
Malaysia +60 1800 80 6789
Netherlands +31800 022 2493 +3120 713 2960
New Zealand +64 800 451650
Norway +47 800 16 836 +47 2193 96 93
Philippines +63 180016510798
Singapore 1800 5792 745
South Korea +82 80 234 5300
Spain +34 800 300 143 +34 911899 417
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
9
Indexed Sequencing Overview Guide
Region Toll Free International
Sweden +46 2 00883979 +46 8 50619671
Switzerland +41800 200 442 +4156 580 00 00
Taiwan, China +886 8 06651752
Thailand +66 1800 011304
United Kingdom +44 800 012 6019 +44 20 7305 7197
United States +1800 809 4566 +1858 202 4566
Vietnam +84 1206 5263
Safety data sheets (SDSs)—Available on the Illumina website at support.illumina.com/sds.html.
Product documentation—Available for download from support.illumina.com.
Document # 15057455 v08
For Research Use Only. Not for use in diagnostic procedures.
10
Indexed Sequencing Overview Guide
