Costa, C.M. and R.P. Roberts. 2014. Techniques for improving the quality and quantity of DNA extracted from herbarium
specimens. Phytoneuron 2014-48: 1–8. Published 12 May 2014. ISSN 2153 733X
TECHNIQUES FOR IMPROVING THE QUALITY AND QUANTITY OF DNA
EXTRACTED FROM HERBARIUM SPECIMENS
CLAYTON M. COSTA and ROLAND P. ROBERTS
Department of Biological Sciences
Towson University
Towson, Maryland 21252
cco[email protected]wson.edu; rroberts@towson.edu
ABSTRACT
There is a need to modify DNA extraction methods to obtain amplifiable DNA from
herbarium specimens that are relatively old –– we explored several methods to improve both quality
and quantity of DNA obtained from a range of specimens of Asteraceae. Leaf tissue was sampled
from herbarium specimens of varying ages. Modifications were made to the Qiagen DNeasy® Plant
Mini Kit and the CTAB Extraction protocols. Comparison of the results of both protocols for recently
collected and older specimens demonstrated that better quality and greater quantity of DNA was
obtained with the modified DNeasy® protocol. The modified DNeasy® protocol was more
consistent in yielding amplifiable DNA from herbarium specimens older than 20 years. Modification
and optimization of two currently used DNA extraction protocols were successful in yielding quality
amplifiable DNA from herbarium specimens of Asteraceae, tribe Astereae, that were up to 127 years
old. The concentration and quality of DNA was comparable to that obtained from specimens less
than 20 years old.
The use of natural history resources in phylogenetics, biogeography, and population biology,
among other areas of biodiversity research, justifies investments in their security and accessibility by
federal, state, and private funding agencies. Among the collections supported by these agencies are
herbaria, repositories of millions of plant specimens, representing documented snapshots of plant
diversity and distribution in space and time. Many have called for increased use of natural history
resources for research into species discovery. It is proposed that large numbers of unknown and/or
undescribed species are part of these collections (Bebber et al. 2010). In addition to prospects of new
species discovery, it is argued that these resources should be utilized in molecular-based studies,
justifying financial and time investments in them (Drábková et al. 2002), while protecting wild
populations from further degradation. However, while many institutions house valuable plant
resources, seldom are researchers able to utilize older plant specimens for molecular investigation. A
major obstacle is the securing of high quality, amplifiable DNA from older herbarium specimens
(Telle & Thines 2008). Molecular studies, particularly among taxa of Asteraceae – the largest family
of flowering plants – can benefit from the use of herbarium resources specifically in cases where taxa
are rare or their distribution remote. Also, use of these resources supports current and future
investments in them and contributes to the conservation and protection of wild populations.
However, despite the abundance of resources in plant collections, researchers are usually restricted to
using more recently collected specimens, those collected within the last 20 years. For older
specimens, the limiting factor is obtaining high quality, amplifiable DNA.
The problems encountered when using older herbarium specimens for molecular studies stem
primarily from specimen collection and processing practices. Post collection processing of
specimens, including alcohol treatment, time between collection and drying, and method of drying
can have adverse effects on the quantity and quality of DNA obtained (Staats et al. 2011). In addition,
the age of the specimens (time since collection) may impede DNA retrieval due to natural DNA
degradation over time (Staats et al. 2011). This combination of natural DNA degradation and post
collection processing practices that accelerate DNA degradation compounds the problem encountered
Costa and Roberts: DNA extraction from herbarium specimens
2
when older plant specimens are used. Damaged or degraded DNA hinders PCR-based molecular
studies and next-generation sequencing (Lindahl 1993). Obtaining quality DNA from old herbarium
specimens usually require several modifications to commonly used protocols (Drábková et al. 2002
and citations therein). Drábková et al. (2002) recommended the use of Qiagen DNeasy® Plant Kit or
the extraction methods from Doyle and Doyle (1987, 1990) as the most satisfactory extraction
methods, albeit for specimens of graminoids only. The resulting extracts from these protocols must
also be purified before they are used as PCR template. These extracts did not, however, consistently
produce amplicons that were larger than 350 bp. Because DNA extraction requires the destructive
sampling of preserved plant specimens, it is imperative that the extraction techniques employed result
in high quality amplifiable DNA, reducing the frequency of sample removal from specimens, thus
preserving the specimens for posterity. In addition to the challenges outlined above, products of plant
secondary chemistry, concentrated by the drying of plant tissues, may have a greater impact on the
quality of DNA recovered from older herbarium specimens. These secondary compounds can impede
downstream use of extracted DNA as they inhibit reactions and processes in which the DNA extract is
utilized.
Here we address some of the issues associated with obtaining high quality, amplifiable DNA
from older herbarium specimens of Asteraceae taxa. We discuss modifications to two commonly
used extraction protocols and their utility among herbarium specimens of varying ages. The
modifications employed are relatively cheap and easy to implement, without substantial increases in
processing time. Specifically, we utilized strategies to increase the concentration of DNA while
reducing the levels of impurities in the DNA extracted from herbarium specimens that are older than
20 years.
Materials and methods
Plant samples for this study were obtained from herbarium specimens. Sampled taxa
included species from Asteraceae, tribe Astereae (Appendix 1). The collection year of specimens
ranged from 2013 to 1887, 0–127 years old (at the time of this study). Two methods of DNA
extraction were explored and modified several times to optimize for DNA quantity and quality.
Samples of desiccated leaves collected close to the apex of specimens were stored in silica gel prior to
DNA extraction. For both methods of extraction, 20-40 mg of leaf tissue was pulverized with a Mini-
Beadbeater-8 (Bio Spec Products Inc. Bartlesville, OK).
Optimization of the hexadecyltrimethylammonium bromide (CTAB) method (modified from
Doyle and Doyle 1987 & 1990) involved reduction of the volume of all reagents to facilitate the use
of micro-centrifuge tubes, a useful though not novel strategy. To the pulverized tissue, 600 µL of 2X
CTAB and 20mg polyvinyl polypyrolidone (PVP) were added and mixed by vortexing. The samples
were incubated for 1 hr 30 min at 70°C, ensuring a more thorough lysis of cells to increase quantity of
DNA obtained from the tissue. Following incubation, 600 µL chloroform:isoamyl alcohol (24:1) was
added to the samples and mixed. Samples were centrifuged for 20 min at 13,000 RPM. The top
aqueous phase was collected for DNA isolation while discarding pellets with cellular debris. In order
to degrade RNA, 2 µL of RNaseA (New England BioLabs Inc., Ipswich, MA) was added to samples
and incubated for 30 min at 37°C. DNA from samples was precipitated with 540 µL of isopropanol (-
20°C), incubated for 15 min at 25°C, then centrifuged for 4 min at 13,000 RPM. Pelleted DNA was
washed with 500 µL of 75% ethanol, centrifuged for 4 min at 13,000 RPM and dried for 5 min in a
Savant SpeedVac® (Thermo Fisher Scientific Inc., Maddison, WI). Dried pellets were resuspended
in 80 µL 1X TE buffer (10mM Tris pH 7.4, 1mM EDTA). To this, 8 µL of 7.5M ammonium acetate
and 180 µL of 100% ethanol were added and mixed. This mixture was incubated for 30 min at 25°C
then centrifuged for 4 min at 13,000 RPM. Pellets were isolated and washed with 500 µL of 75%
ethanol, centrifuged for 4 min at 13,000 RPM, then dried in a SpeedVac® for 5 min. The pelleted-
DNA was resuspended in 100 µL 1X TE and stored at -20°C.
Costa and Roberts: DNA extraction from herbarium specimens
3
DNA was also extracted from pulverized leaf tissue using the Qiagen DNeasy® Plant Mini
Kit and protocol. The protocol was modified and optimized for extraction of DNA from recently
collected and ancient herbarium specimens. Pulverized tissue was mixed with 450 µL of Buffer AP1,
4 µL of RNaseA and mixed vigorously by vortexing. The mixture was then incubated for 60 min at
70°C to lyse the cells. After lysing, 130 µL of Buffer AP2 was added and the lysate incubated for 60
min at 4°C to precipitate detergent, proteins and polysaccharides. After this precipitation step, the
DNeasy® protocol was followed as prescribed by the manufacturer until the DNA elution step. To
elute the DNA 50 µL of Buffer AE was added to the spin column followed by an extended incubation
time of 10 min, instead of the recommended 5 min. This step was repeated to reach final volume of
100µL.
DNA quality and concentration were evaluated using a NanoDrop Lite (NanoDrop Products,
Wilmington, Delaware). We recorded A260/A280 ratios as indicative of DNA purity (1.7-1.9
indicating pure DNA) and the concentration of DNA in each sample. Extracted DNA samples were
stored as stock solutions at -20°C. Subsequently, working solutions were prepared through serial
dilutions for use as template in polymerase chain reactions (PCR). PCR utilized nuclear ribosomal
DNA (nrDNA), spanning external transcribed spacer (ETS) and internal transcribed spacer (ITS) and
chloroplast DNA, spanning psbA-trnH and ycf1 3300-4280 for amplification of DNA. In order to
access the effectiveness of these extracts in amplifying DNA we used regions of variable lengths,
350-550 bp in ETS, 600-900 bp in ITS, 250-400 bp in psbA-trnH and 700-1000 bp in ycf1 3300-4280.
Results and Discussion
Extracts obtained using the unmodified CTAB protocol produced high concentrations of
DNA, on average 234 ng/µL (mean A260/A280 ratio = 1.63). However, we were unable to obtain
amplicons with these extracts when used as PCR template. The same taxa were sampled using the
unmodified DNeasy® protocol, which produced on average 92.5 ng/µL (mean A260/A280 ratio =
1.24). These extracts also failed to produce amplicons for most DNA regions tested. Extracts
obtained from modifications to the CTAB protocol displayed 519.8 ng/µL mean DNA concentration
and mean A260/A280 ratio of 1.32. When the samples were subjected to the modified DNeasy®
protocol the DNA concentration of the extracts was on average 87.9 ng/µL (mean A260/A280 ratio =
1.38). Comparisons of extracts obtained from the modified DNeasy® and CTAB protocols showed
that extracts obtained from the modified DNeasy® protocol were more consistent in amplicon
production when used as PCR template. This was the case despite the tendency of the modified
CTAB protocol to yield higher concentrations of DNA compared to the modified DNeasy® protocol.
As a result, we continued to refine and optimize the DNeasy® protocol for extraction of DNA from
older herbarium specimens, resulting in amplifiable DNA from specimens collected 127 years ago
(Table 1).
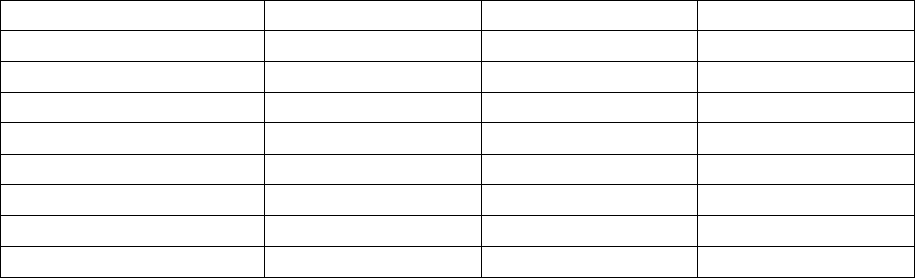
Table 1. DNA quality and quantity using the modified DNeasy® method on recent and old herbarium
specimens.
Taxon Name Year collected A260/A280 [DNA] (ng/µL)
Chrysopsis mariana 1967 1.65 71.1
Chrysopsis mariana 2008 1.65 79.6
Pityopsis ruthii 1900 1.40 44.8
Pityopsis ruthii 2013 1.46 55.0
Heterotheca viscida 1963 1.48 37.9
Heterotheca viscida 2000 1.44 16.4
Noticastrum marginatum 1977 1.51 9.0
Noticastrum marginatum 2011 1.25 94.6
Costa and Roberts: DNA extraction from herbarium specimens
4
The optimized DNeasy® protocol consistently resulted in amplifiable DNA in more cases
than the CTAB protocol. In particular, regions of chloroplast DNA extracted using the modified
DNeasy® protocol could be amplified from specimens that were over 100 years old. We recovered,
on average, 60 ng/µL of DNA (mean A260/A280 ratio = 1.54) using the modified DNeasy® protocol
(Table 2). The total time required for this method increased to approximately 3 hr compared to the
original 1 hr. We recovered an average of 406 ng/µL of DNA (mean A260/A280 ratio = 1.4) using
the modified CTAB method, and decreased time required for this protocol from ~8 hr to <4 hr (Table
1). The quality of DNA extracted using the modified DNeasy® protocol were significantly better
than those from the modified CTAB extractions (p=0.048), where seven of the nine sampled taxa
produced DNA of higher purity (Table 2).
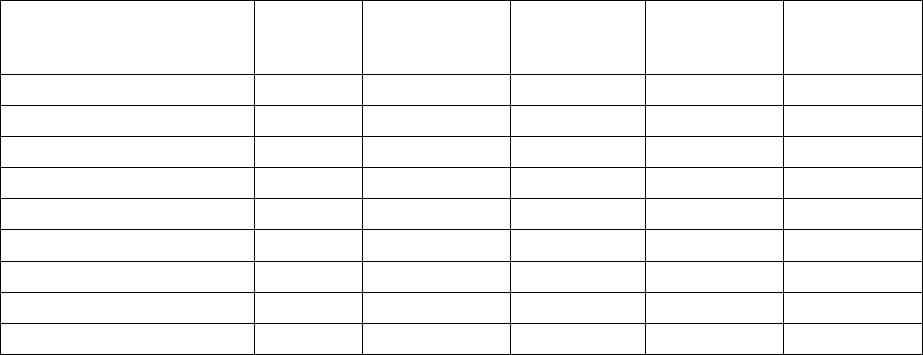
Table 2. DNA quality (spectrophotometer A260/A280 where 1.70-1.9 indicates pure DNA) and quantity using a
modified CTAB protocol and modified DNeasy® method. Note:
f
Extracts from modified CTAB failed to
amplify when used as template in PCR for most genes.
Taxon Name
Year
collected
CTAB
A260/A280
QIAGEN
A260/A280
CTAB
[DNA]
(ng/µL)
QIAGEN
[DNA]
(ng/µL)
Chrysopsis mariana
f
1967 1.50 1.65 417.0 71.1
Chrysopsis gossypina 1974 1.27 1.71 188.5 70.1
Heterotheca oregona 1986 1.43 1.48 466.7 41.3
Croptilon divaricatum 1987 1.42 1.43 133.0 25.3
Tomentaurum niveum 1929 1.57 1.65 702.3 59.0
Tomentaurum niveum
f
1887 1.30 1.14 414.7 14.8
Heterotheca inuloides
f
1970 1.35 1.53 470.9 66.8
Biglowia nudata 2000 1.19 1.56 161.1 47.5
Euthamia graminifolia 2001 1.58 1.73 705.1 150.1
The potential benefits of the extracting DNA from old herbarium specimens are great, as this
adds to the value of millions of specimens currently housed in natural history collections. We have
increased both the quantity and quality of DNA extracted from herbarium specimens of Asteraceae
dating up to 127 years old, a large improvement when compared to the oldest specimen, 71 years, in
Drábková et al. (2002). We increased DNA concentration using DNeasy® Plant Mini Kit by
modifying lysis time for cells, the volume of Buffer AP1 used per reaction, and increased incubation
time prior to DNA elution. These modifications also resulted in increased DNA purity (Table 2).
Although the overall time required for this protocol has increased, we believe that the yield of
amplifiable DNA from relatively old herbarium specimens outweighs the increase in extraction time.
While Drábková et al. (2002) suggested their methods as useful for specimens of graminoids,
we have demonstrated a wider application of these extraction methods for specimens of Asteraceae,
which have vastly different secondary chemistry and anatomy. We have also reduced the amount of
material needed for extractions (between 20-40 mg instead of 100-500 mg). Additionally, DNA
extracts from the same specimen or related taxa, ranging from 0 to 127 years old, were used to
evaluate the efficacy of the modified and optimized DNeasy® protocol. Interestingly, unlike findings
in Drábková et al. (2002), our extracts resulted in successful amplification of DNA from both nuclear
and chloroplast regions, ranging from 350 to 1000 bp. These modified protocols were also successful
when used for recently collected specimens, increasing the range of specimens that can be harnessed
for molecular studies.

Costa and Roberts: DNA extraction from herbarium specimens
5
ACKNOWLEDGEMENTS
We thank the Department of Biological Sciences at Towson University. We wish to thank
herbaria at Delaware State University (DOV), University of Texas, Austin (TEX/LL), and the Smithsonian
Institution (US) for specimens. In addition, we thank Dr. Edward Schilling and Dr. John C. Semple for
specimens used in this study.
LITERATURE CITED
Bebber, D.P., M.A. Carine, J.R. Wood, A.H. Wortley, D.J. Harris, G.T. Prance, G. Davidse, J. Paige, T.D.
Pennington, N.K. Robson, and R.W. Scotland. 2010. Herbaria are a major frontier for species
discovery. Proc. Natl. Acad. Sci. USA 107: 22169–22171.
Doyle, J.J., and J.L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf
tissue. Phytochem. Bull. 19: 11–15.
Doyle, J. J. and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15.
Drábková, L., J. Kirschner, and Č. Vlček. 2002. Comparison of seven DNA extraction and amplification
protocols in historical herbarium specimens of Juncaceae. Pl. Molec. Biol. Rep. 20: 161–175.
Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715.
Telle, S. and M. Thines. 2008. Amplification of cox2 (~620 bp) from 2 mg of up to 129 years old
herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLOS ONE 3:
e3584.
Staats, M., A. Cuenca, J.E. Richardson, R.V. Ginkel, G. Petersen, O. Seberg, and F.T. Bakker. 2011. DNA
damage in plant herbarium tissue. PLOS ONE 6: e28448.
APPENDIX 1. Voucher information for specimens used in this study
Taxon Name
Collector and number
Collection
Date
Collection Locality Herbarium
Bigelowia nudata Kral 90535 2000 AL: Washington Co. DOV
Bradburia hirtella McCrarry 72 1971 TX: Nacogdoches Co. BALT
Bradburia hirtella Jones & Jones 455 1987 TX: Brazos Co. US
Bradburia pilosa
Allen 8361 & Vincent
1685 1978 LA: Union Parish BALT
Chrysopsis gossypina Pittillo 2807 1965 GA: Burke Co. BALT
Chrysopsis gossypina Rothwell C-60 1974 NC: Jones Co. BALT
Chrysopsis linearifolia Davis & Davis 15565 1970 FL: Walton Co. BALT
Chrysopsis mariana D'Arcy 2219 1967 FL: Alachua Co. BALT
Chrysopsis mariana Strong 3787 2008 FL: Marion Co. US
Euthamia graminifolia Naczi 8917 2001 DE: Kent Co. DOV
Heterotheca inuloides var. rosei
Cronquist & Fay
10815 1970 Mexico US
Heterotheca oregona var.
scaberrima Semple & Heard 8588 1986 CA: Santa Clara Co. BALT
Heterotheca viscida Bennett 8306 1963 NM: Santa Fe Co. US
Heterotheca viscida Carr & Karges 19141 2000 TX: Jeff Davis Co. TEX/LL
Noticastrum marginatum
Norrbom et al. 11-PE-
17
2011 Peru US
Noticastrum marginatum Olsen & Escobar 576 1977 Colombia TEX/LL
Pityopsis ruthii Ruth 1501 1900 TN: Hiawassee Valley US
Pityopsis ruthii Schilling (Aug. 2013) 2013 TN: Polk Co. TENN
Tomentaurum niveum Mexia 2598 1929 Mexico US
Tomentaurum niveum Pringle s.n. 1887 Mexico US
Costa and Roberts: DNA extraction from herbarium specimens
6
APPENDIX 2. CTAB extraction protocol
Sample Preparation
• Prepare 2ml tubes for tissue homogenizer with labels and white zirconia/silica beads
• Warm 2X CTAB to 60°C in water bath
• Homogenize samples in Mini Bead Beater 8 for 2min
• Spin for 1min at 13,000 RPM
• Under the hood
o Add 600µl 2X CTAB to homogenized sample
o Add small volume of PVP with sterilized spatula
• Vortex samples; invert and vortex again
Extraction and Isolation of DNA
• Incubate for 1hr 30min at 70°C.
o Mix manually (DO NOT VORTEX) every 30min
• Add 600µl 24:1 chloroform: isoamyl alcohol to sample
o Manually mix samples
• Spin for 20min at 13,000 RPM
o Check for clear liquid and spin again if necessary
• Under the hood
o Transfer top aqueous phase only to a new 1.5ml microfuge tube
DNA Precipitation
• Add 2µL of RNaseA and incubate 30min at 37°C
• Add 540µl cold isopropanol (-20°C)
o Mix manually
• Incubate for 15min at 25°C
• Spin samples for 4min at 13,000 RPM
o Decant supernatant (DNA is present in the pellet)
• Wash pellet with 500µl 75% EtOH and invert tubes or flick w/ finger several times
• Spin samples for 4min at 13,000 RPM
o Decant supernatant and dry excess liquid by blotting with Kimwipe
Costa and Roberts: DNA extraction from herbarium specimens
7
• Dry with SpeedVac for 5min
Final Cleaning and Re-Suspension of DNA
• Resuspend pellet in 80µl 1X TE; break up pellet completely
• Add 8µl of 7.5M ammonium acetate, and 180µl of 100% EtOH
• Mix manually and incubate for 30min at 25°C
• Spin for 4min at 13,000 RPM, decant supernatant
• Add final wash of 500µl 75% EtOH, gently invert tubes
• Spin 4min at 13,000 RPM
o Decant supernatant and dry excess liquid by blotting with Kimwipe
• Dry with SpeedVac for 5min
• Resuspend pelleted DNA with 100µL 1X TE and store at -20°C
APPENDIX 3. QIAGEN DNeasy® Plant Kit Protocol
• Disrupt samples (20-40mg desiccated plant tissue) using Mini Beadbeater-8
o Centrifuge samples @ 14,000 RPM for 1 min
• Add 450µL Buffer AP1 and 4µL RNase A
o Vortex samples to thoroughly mix
o Incubate at 70°C for 60 min
o Transfer to NEW 2mL microfuge tube
• Add 130µL Buffer P3
o Mix by inversion
o Incubate in ice for 60 min
• Centrifuge the lysate @ 14,000 RPM for 5 min
• Pipet clear lysate into a QIAshredder spin column (lilac) placed in a 2mL collection tube
o Centrifuge @ 14,000 RPM for 2 min
o Transfer the flow-through fraction (measure volume) into NEW 1.5mL microfuge
tube without disturbing the cell-debris pellet
• Add 1.5 volumes of Buffer AW1. MIX IMMEDIATELY
Costa and Roberts: DNA extraction from herbarium specimens
8
• Pipet 650µL of the mixture into DNeasy® Mini spin column (white) placed in a 2mL
collection tube
o Centrifuge @ 8,000 RPM for 1 min
o Discard flow-through. Blot-dry the collection tube and reuse
• Repeat step 7 with remaining sample
o Centrifuge @ 8,000 RPM for 1 min
o Discard flow-through and collection tube
o Place DNeasy® Mini spin column into NEW 2mL collection tube
• Add 500µL Buffer AW2
o Centrifuge @ 8,000 RPM for 1 min
o Discard flow-through. Blot-dry the collection tube and reuse
• Add 500µL Buffer AW2
o Centrifuge @ 14,000 RPM for 2 min
• Remove the spin column from collection tube carefully so that the column does not come into
contact with the flow-through
o Discard flow-through and collection tube
• Transfer the spin column to a new 1.5mL microfuge tube
• Add 50µL Buffer AE for elution
o Incubate at room temperature for 10 min
o Centrifuge @ 8,000 RPM for 1 min
• Repeat previous step.
• Store in -20°C
