GE Healthcare
Life Sciences
Biacore™ Assay Handbook


Biacore Assay Handbook 29-0194-00 Edition AA 1
1 Introduction
1.1 What Biacore™ systems measure .............................................. 5
1.2 Where Biacore systems are used ................................................ 5
1.3 Scope of this book .......................................................................... 6
1.4 Biacore system range ................................................................... 6
1.5 Biacore terminology ...................................................................... 7
2 Application overview
2.1 Screening and detecting binding partners ............................... 9
2.2 Kinetics and affinity measurements ........................................ 10
2.2.1 Kinetic analysis...........................................................................................10
2.2.2 Affinity analysis..........................................................................................11
2.3 Concentration measurements .................................................. 13
2.3.1 Calibrated assays......................................................................................13
2.3.2 Calibration-free concentration analysis (CFCA)............................14
2.4 Mapping binding sites ................................................................. 14
3 General considerations
3.1 Sensor surfaces ............................................................................ 17
3.1.1 General properties....................................................................................17
3.1.2 Sensor surface types................................................................................17
3.2 Attaching the ligand .................................................................... 19
3.2.1 Covalent immobilization ........................................................................19
3.2.2 High affinity capture ................................................................................21
3.2.3 Response levels..........................................................................................21
3.3 General buffer considerations ................................................... 23
3.3.1 Buffer substances......................................................................................23
3.3.2 Ionic strength ..............................................................................................23
3.3.3 Additives........................................................................................................24
3.3.4 Buffer preparation ....................................................................................25
3.4 Matching sample and running buffer ...................................... 26
3.4.1 Matching refractive index......................................................................26
3.4.2 Matching buffer composition...............................................................27
3.4.3 Matching buffer environment in practice.......................................28
3.5 Dilution series and replicates .................................................... 28
3.5.1 Dilution series..............................................................................................28
3.5.2 Replicates .....................................................................................................29
3.6 Preparing vials and microplates ............................................... 30
4 Screening and detecting binding partners
4.1 Small molecule screening ........................................................... 32
4.1.1 Goals...............................................................................................................32
4.1.2 Sensor surface preparation..................................................................32
4.1.3 Sample preparation .................................................................................32
4.1.4 Buffers............................................................................................................32

2 Biacore Assay Handbook 29-0194-00 Edition AA
4.1.5 Analysis conditions...................................................................................33
4.1.6 Evaluating results......................................................................................34
4.2 Antibody screening ......................................................................35
4.2.1 Goals...............................................................................................................35
4.2.2 Sensor surface preparation..................................................................36
4.2.3 Sample preparation .................................................................................36
4.2.4 Buffers............................................................................................................36
4.2.5 Analysis conditions...................................................................................36
4.2.6 Evaluating results......................................................................................37
4.3 Immunogenicity testing ..............................................................38
4.3.1 Goals and challenges..............................................................................38
5 Kinetics and affinity measurements
5.1 Approaches ....................................................................................39
5.1.1 Single- and multi-cycle kinetics...........................................................39
5.1.2 2-over-2 kinetics........................................................................................39
5.1.3 Comparative estimates of kinetics and affinity............................40
5.2 Sensor surface preparation ........................................................40
5.3 Buffers ............................................................................................41
5.4 Sample preparation .....................................................................41
5.5 Sample concentrations ...............................................................42
6 Epitope mapping
6.1 Pair-wise binding ..........................................................................45
6.1.1 Principle.........................................................................................................45
6.1.2 Evaluation.....................................................................................................46
6.2 Peptide inhibition .........................................................................48
7 Troubleshooting assays
7.1 Troubleshooting ligand attachment .........................................49
7.1.1 Covalently attached ligand...................................................................49
7.1.2 Captured ligand .........................................................................................50
7.2 Troubleshooting analyte binding ..............................................51
7.2.1 Analyte capacity too low .......................................................................51
7.2.2 Analyte binding too high........................................................................51
7.3 Dealing with non-specific and unwanted binding .................51
7.3.1 Non-specific binding................................................................................51
7.3.2 Unwanted binding....................................................................................52
7.4 Unexpected sensorgram shapes ...............................................52
7.4.1 Unstable baseline......................................................................................53
7.4.2 Sample response below baseline.......................................................54
7.4.3 “Humpbacked” sensorgrams...............................................................55
7.4.4 Anomalous response during buffer flow ........................................55
7.4.5 Response subtraction spikes................................................................56
7.4.6 Regular disturbances ..............................................................................56

Biacore Assay Handbook 29-0194-00 Edition AA 3
Appendix A Analysis of kinetics and concentration measurements
A.1 Basic principles of kinetics and affinity ................................... 57
A.1.1 Kinetic rate equations .............................................................................57
A.1.2 Steady-state affinity equations...........................................................59
A.1.3 Fitting procedure.......................................................................................59
A.1.4 Assessing the fit .........................................................................................60
A.2 Interaction models for kinetics ................................................. 62
A.2.1 1:1 binding....................................................................................................62
A.2.2 1:1 dissociation ..........................................................................................63
A.2.3 Bivalent analyte binding ........................................................................63
A.2.4 Heterogeneous ligand.............................................................................64
A.2.5 Interaction models for affinity .............................................................64
A.3 Concentration measurements .................................................. 64
A.3.1 Calibration curve fitting..........................................................................64
A.3.2 Calibration trends......................................................................................65
A.3.3 Calibration-free concentration analysis..........................................65
Appendix B Solvent correction principles and practice
B.1 Introduction .................................................................................. 67
B.2 Requirement for solvent correction ......................................... 67
B.3 Solvent correction principles ..................................................... 68
B.4 Preparing solutions for solvent correction ............................. 69
B.5 Assessing solvent correction procedures ............................... 69

4 Biacore Assay Handbook 29-0194-00 Edition AA

Biacore Assay Handbook 29-0194-00 Edition AA 5
Introduction 1
1 Introduction
1.1 What Biacore™ systems measure
Biacore systems monitor molecular interactions in real time, using a non-
invasive label-free technology that responds to changes in the concentration of
molecules at a sensor surface as molecules bind to or dissociate from the
surface. The detection principle is based on surface plasmon resonance (SPR),
that is sensitive to changes in refractive index within about 150 nm from the
sensor surface. To study the interaction between two binding partners, one
partner is attached to the surface and the other is passed over the surface in a
continuous flow of sample solution. The SPR response is directly proportional to
the change in mass concentration close to the surface.
Biacore systems can be used to study interactions involving (in principle) any
kind of molecule, from organic drug candidates to proteins, nucleic acids,
glycoproteins and even viruses and whole cells. Since the response is a measure
of the change in mass concentration, the response per molar unit of interactant
is proportional to the molecular weight (smaller molecules give lower molar
responses). The practical lower limit for detection of small molecules with
today’s instrumentation is about 100 Da.
The detection principle does not require any of the interactants to be labeled,
and measurements can be performed on complex mixtures such as cell culture
supernatants or cell extracts as well as purified interactants. The identity of the
interactant monitored in a complex sample matrix is determined by the
interaction specificity of the partner attached to the surface. The SPR detection
principle is non-invasive and works equally well on clear and colored or opaque
samples.
1.2 Where Biacore systems are used
Measurements with Biacore systems provide valuable information in a number
of application areas:
• detection of specific molecules such as anti-drug antibodies in
immunogenicity studies
• screening for binding partners and ranking of binding ability, for instance
in drug discovery and biopharmaceutical development
• determination of simultaneous interaction capabilities, for example
epitope mapping with monoclonal antibodies

1 Introduction
1.3 Scope of this book
6 Biacore Assay Handbook 29-0194-00 Edition AA
• determination of interactant concentration in samples, from
measurements under conditions where interaction levels can be related to
concentration
• measurement of interaction kinetics and affinity, made possible by real-
time detection of interaction events
Biacore systems are used primarily in pharmaceutical development, quality
control and basic life science research.
1.3 Scope of this book
This book gives a general overview of the different types of Biacore-based
applications, together with advice and recommendations on preparation of the
sensor surface, preparation of samples and design and optimization of different
assays. Troubleshooting guidelines to help in interpreting unsatisfactory results
are also included.
The information is presented in general terms, without reference to specific
Biacore systems. Use this handbook together with the specific user
documentation for your Biacore system, which will describe how to implement
the assay principles in the instrument and system software.
1.4 Biacore system range
All Biacore systems exploit the same detection principle and use essentially the
same range of sensor surfaces (see Section 3.1), with a controlled flow system
for delivery of samples and reagents to the sensor surface. Individual systems
differ primarily in capacity and degree of automation, and are suited to use in
different laboratory contexts:
• Entry-level systems like Biacore X100 have one or two pairs of flow cells
(where one cell in a pair is used for detecting the interaction and the other
as a reference cell where required) and can handle only relatively few
samples without user intervention.
• Advanced systems may have multiple detection spots (in Biacore 4000, for
example, 20 detection spots are divided between 4 flow cells for parallel
analysis of multiple samples) and unattended sample handling capacity
for 1500 samples or more.
While the available options for assay development and execution may vary
between the different systems, the same general principles apply to all systems.
This book will focus as far as possible on common principles of Biacore assays,
keeping system-specific information to a minimum.
Biacore Assay Handbook 29-0194-00 Edition AA 7
Introduction 1
1.5 Biacore terminology
Biacore systems monitor the interaction between two molecules, of which one
is attached to the sensor surface and the other is free in solution. The following
terms are used in the context of Biacore-based assays:
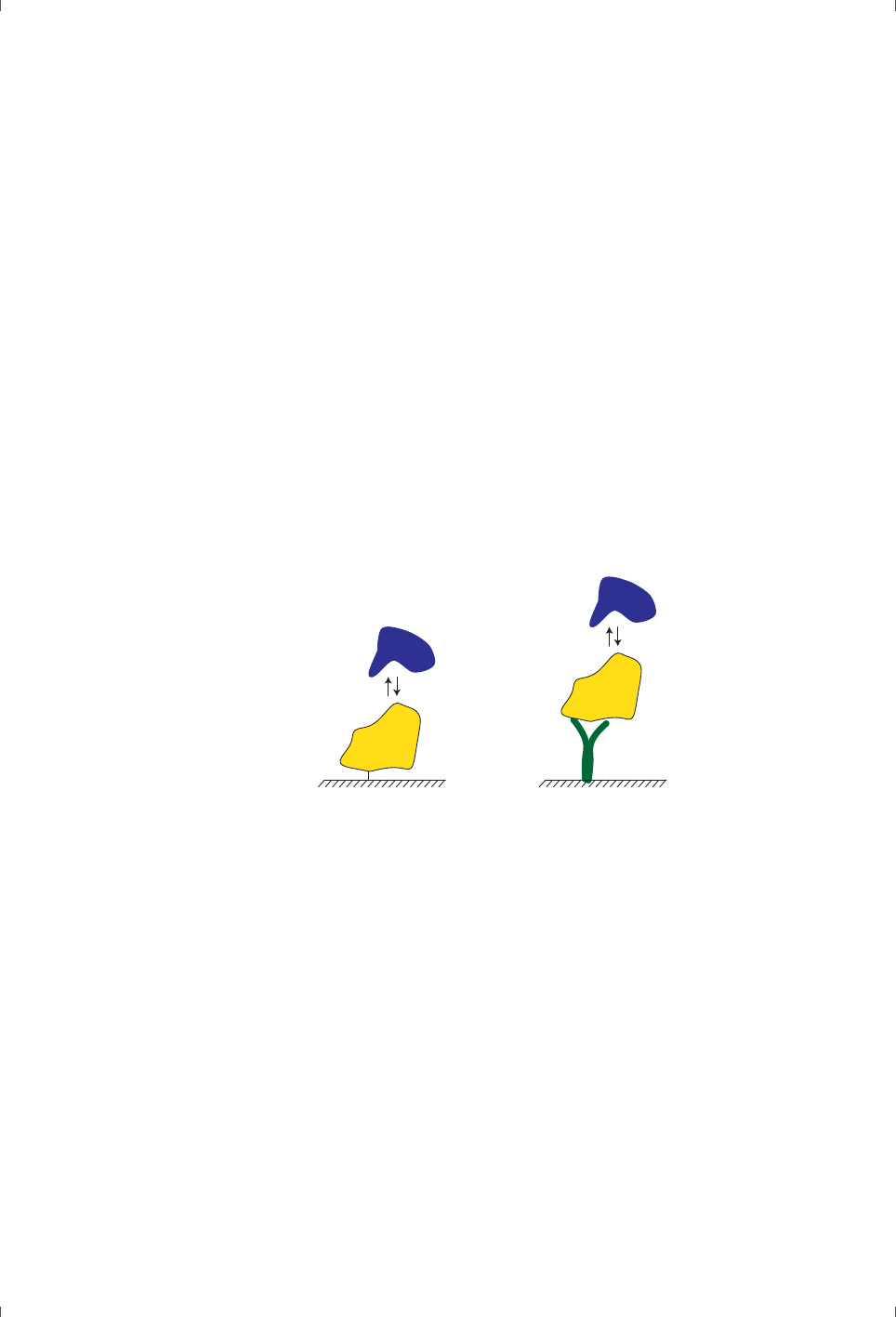
• The interaction partner attached to the surface is called the ligand (Figure
1-1). In drug discovery and development work, the ligand is sometimes
referred to as the target molecule.
Note: The term “ligand” is applied here in analogy with terminology used in
affinity chromatography contexts, and does not imply that the
surface-attached molecule is a ligand for a cellular receptor.
• The ligand may be attached to the surface either by covalent
immobilization using chemical coupling reagents or by capturing through
high affinity binding to an immobilized capturing molecule.
•The analyte is the interaction partner that is passed in solution over the
ligand (Figure 1-1).
Figure 1-1. The ligand is the interaction partner that is attached to the sensor
surface. The ligand may be immobilized directly on the surface (left) or attached
through binding to an immobilized capturing molecule (right). The analyte is free in
solution and binds to the immobilized ligand.
• In indirect assay formats the analyte is detected using a secondary
detecting molecule, which can bind to both analyte in solution and ligand
on the sensor surface. The observed response is derived from binding of
detecting molecule to the ligand: the presence of analyte in the sample
inhibits this binding so that the response is inversely related to the amount
of analyte.
• Analysis is performed by injecting sample over the surface in a carefully
controlled fashion. The sample is carried in a continuous flow of buffer,
termed running buffer.
• Response is measured in resonance units (RU). The response is directly
proportional to the concentration of biomolecules on the surface.
ligand
analyte
sensor
surface
ligand
analyte
capturing
molecule
1 Introduction
1.5 Biacore terminology
8 Biacore Assay Handbook 29-0194-00 Edition AA
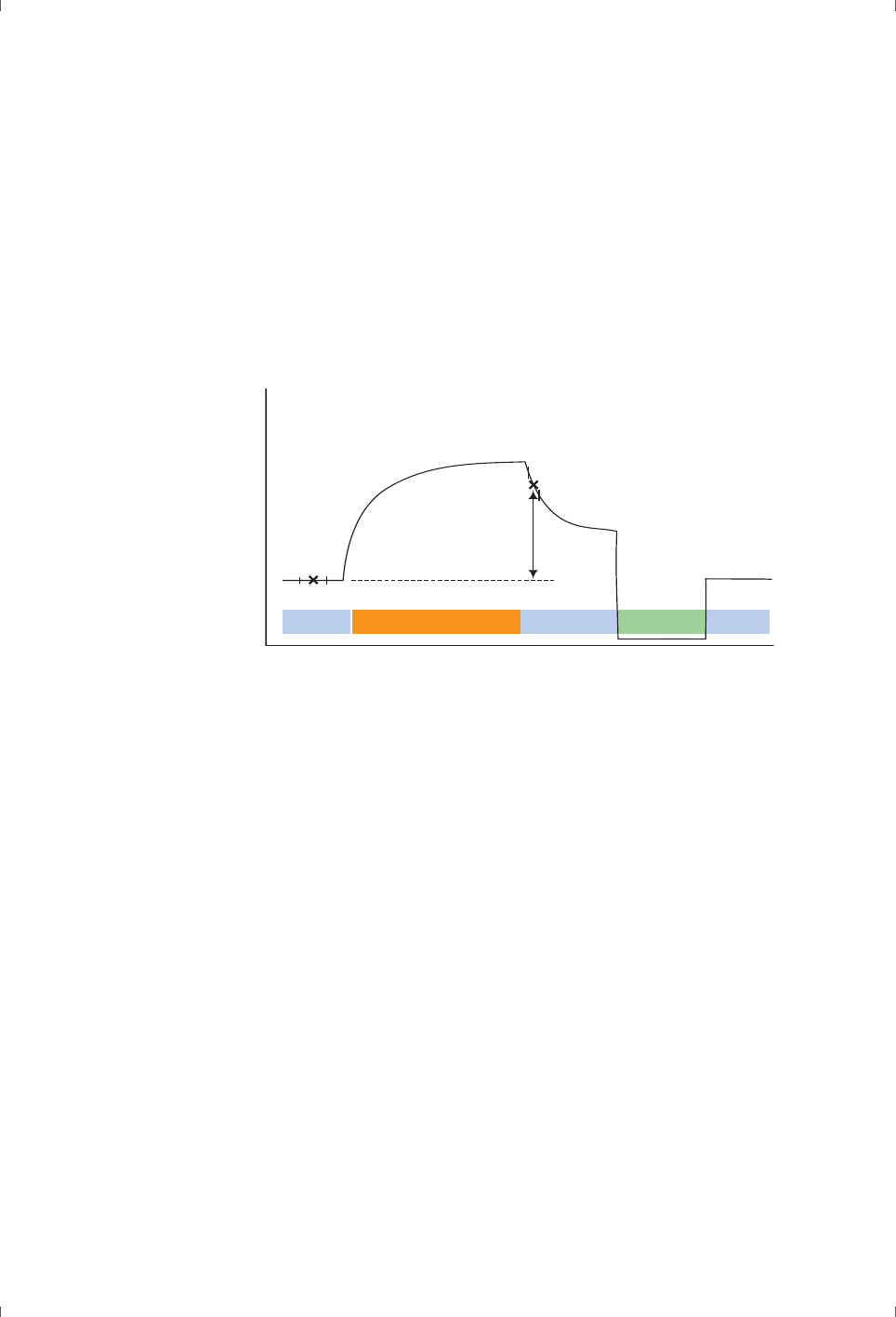
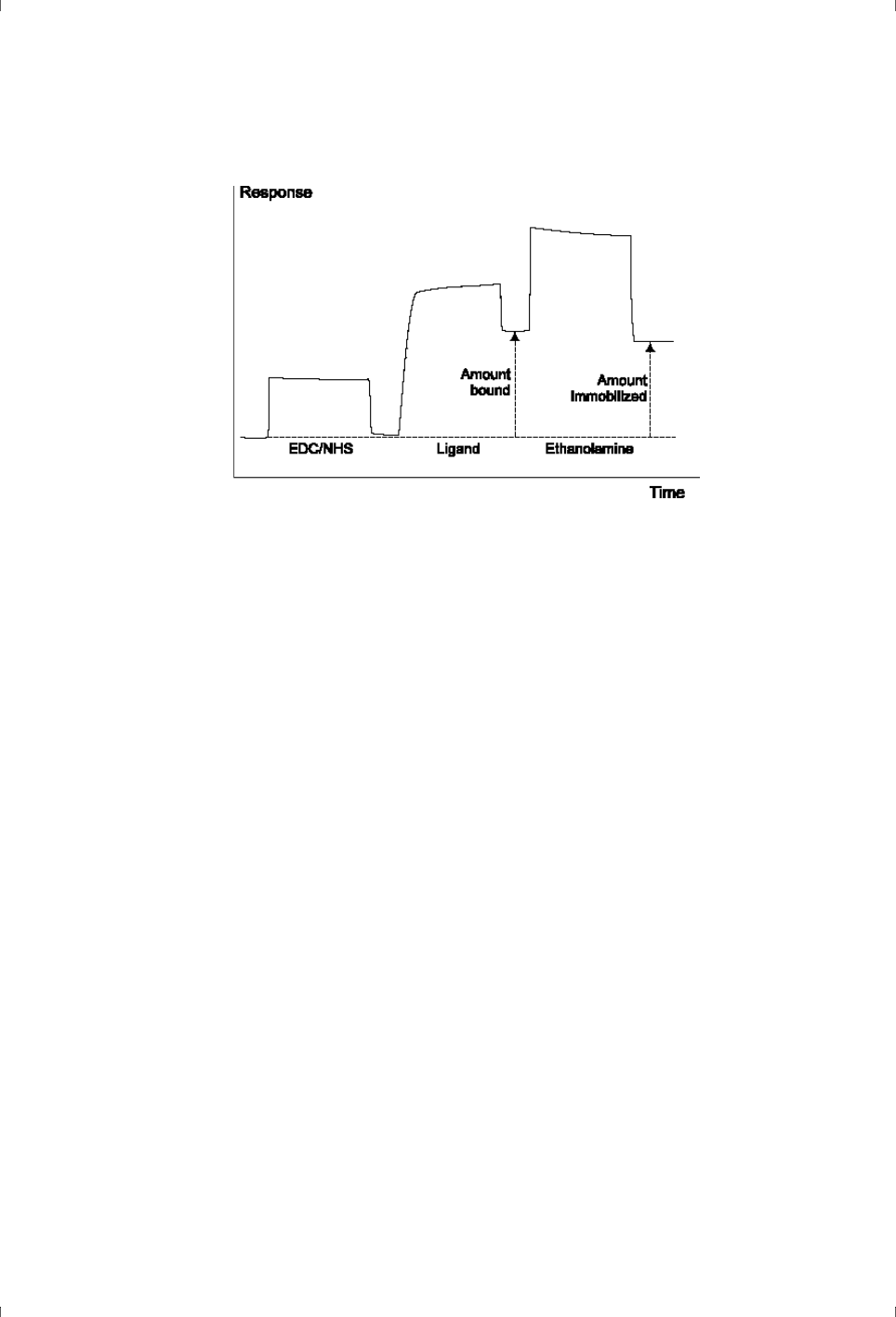
•A sensorgram is a plot of response against time, showing the progress of
the interaction (Figure 1-2). This curve is displayed directly on the
computer screen during the course of an analysis.
•A report point records the response on a sensorgram at a specific time
averaged over a short time window, as well as the slope of the sensorgram
over the window. The response may be absolute (above a fixed zero level
determined by the detector) or relative to the response at another
specified report point (Figure 1-2).
Figure 1-2. Schematic illustration of a sensorgram. The bars below the sensorgram
curve indicate the solutions that pass over the sensor surface.
• Regeneration is the process of removing bound analyte from the surface
after an analysis cycle without damaging the ligand, in preparation for a
new cycle. For ligand that is captured rather than immobilized,
regeneration usually removes the ligand and leaves the capturing
molecule intact.
• SPR detection monitors changes in refractive index close to the surface,
and differences in refractive index between running buffer and injected
sample will be recorded as a rapid shift in response at the beginning and
end of the injection. This is referred to as a bulk refractive index effect or
bulk shift.
• Naming conventions for kinetic and affinity constants vary considerably in
the literature. Throughout this book (and in other Biacore documentation),
rate constants are identified with lower-case letters (k
a
for association rate
constant and k
d
for dissociation rate constant), while equilibrium or affinity
constants are written with upper-case letters (K
A
for the equilibrium
association constant and K
D
for the equilibrium dissociation constant).
Absolute
response (RU)
Report point
(baseline)
Report point
(response)
Relative
response (RU)
Running
buffer
Running
buffer
Running
buffer
Sample
Regeneration
solution
Time

Biacore Assay Handbook 29-0194-00 Edition AA 9
Application overview 2
2 Application overview
This chapter gives an overview of the major application areas for Biacore
systems. These are
• Screening for binding partners, including immunogenicity testing
• Measurement of interaction kinetics and affinity
• Measuring concentration
• Mapping binding site topography
Subsequent chapters in this Handbook consider screening, kinetics/affinity and
binding site topography in more detail. Measurement of concentration is
discussed in a separate Handbook (Biacore Concentration Analysis Handbook).
2.1 Screening and detecting binding partners
Because Biacore systems exploit surface plasmon resonance (SPR), a generic
detection technology that requires no labels, they are well suited to screening
for binding partners in pharmaceutical and biopharmaceutical development
and immunogenicity testing.
The response in a Biacore system is directly related to the change in mass
concentration on the surface, so that molar responses (i.e. responses for a given
number of molecules) are proportional to the size of the molecule involved. A
given response will represent a higher molar concentration of a small molecule
than a large one: conversely, a given number of molecules binding to the
surface will give a lower response if the molecule is small. The relationship
between response and surface concentration is essentially constant for
proteins regardless of amino acid composition and sequence, and is similar for
most other biological macromolecules.
Note: In early literature related to Biacore, 1 RU was stated to be approximately
equivalent to a change in surface concentration of 1 pg/mm
2
. This
relationship holds roughly for proteins on Sensor Chip CM5. Beware of
applying the conversion to non-protein molecules and other types of
sensor chip. Response values from Biacore systems should always be
quoted in RU, not in units of surface concentration.
Measurements can be made on complex samples such as cell culture medium
or body fluids: the specificity of the observed response is determined primarily
by the choice of ligand attached to the surface. This is particularly useful in
applications such as antibody screening and immunogenicity testing: for
example, screening with an antigen as ligand will detect only specific
antibodies, while using an ligand directed towards a common region of a given
antibody class will detect that class regardless of antigen specificity.
2 Application overview
2.2 Kinetics and affinity measurements
10 Biacore Assay Handbook 29-0194-00 Edition AA
2.2 Kinetics and affinity measurements
Determination of interaction kinetics is perhaps the most characteristic
application for Biacore systems. The label-free real-time detection allows
interactions to be monitored with high resolution as they happen, and the
results can be interpreted in relation to a mathematical model of the interaction
mechanism to evaluate kinetic parameters (association and dissociation rate
constants). Affinity constants, which reflect the strength of binding but not the
rate, can be derived either from the rate constants or from analysis of the level
of binding at steady state. The theory of kinetic and affinity analysis is described
in Appendix A.
Note: The term “kinetics” is used in this handbook to refer to interaction kinetics.
The kinetics of other processes such as enzyme-catalyzed reactions are
not normally amenable to study in Biacore systems.
2.2.1 Kinetic analysis
Interaction kinetics are analyzed by monitoring the interaction as a function of
time over a range of analyte concentrations, and then fitting the whole data set
to a mathematical model describing the interaction. The association phase
(during sample injection) contains information on both association and
dissociation processes, while only dissociation occurs during the dissociation
phase (after sample injection, when buffer flow removes dissociated analyte
molecules).
It is important to be aware that the results of kinetic analysis are relevant only
in the context of the interaction model chosen for the evaluation. It is not strictly
possible to derive a model from the observed binding behavior, although the
shape of the sensorgrams can sometimes give clues for the choice of
appropriate models. Fitting the data to a mathematical model does not provide
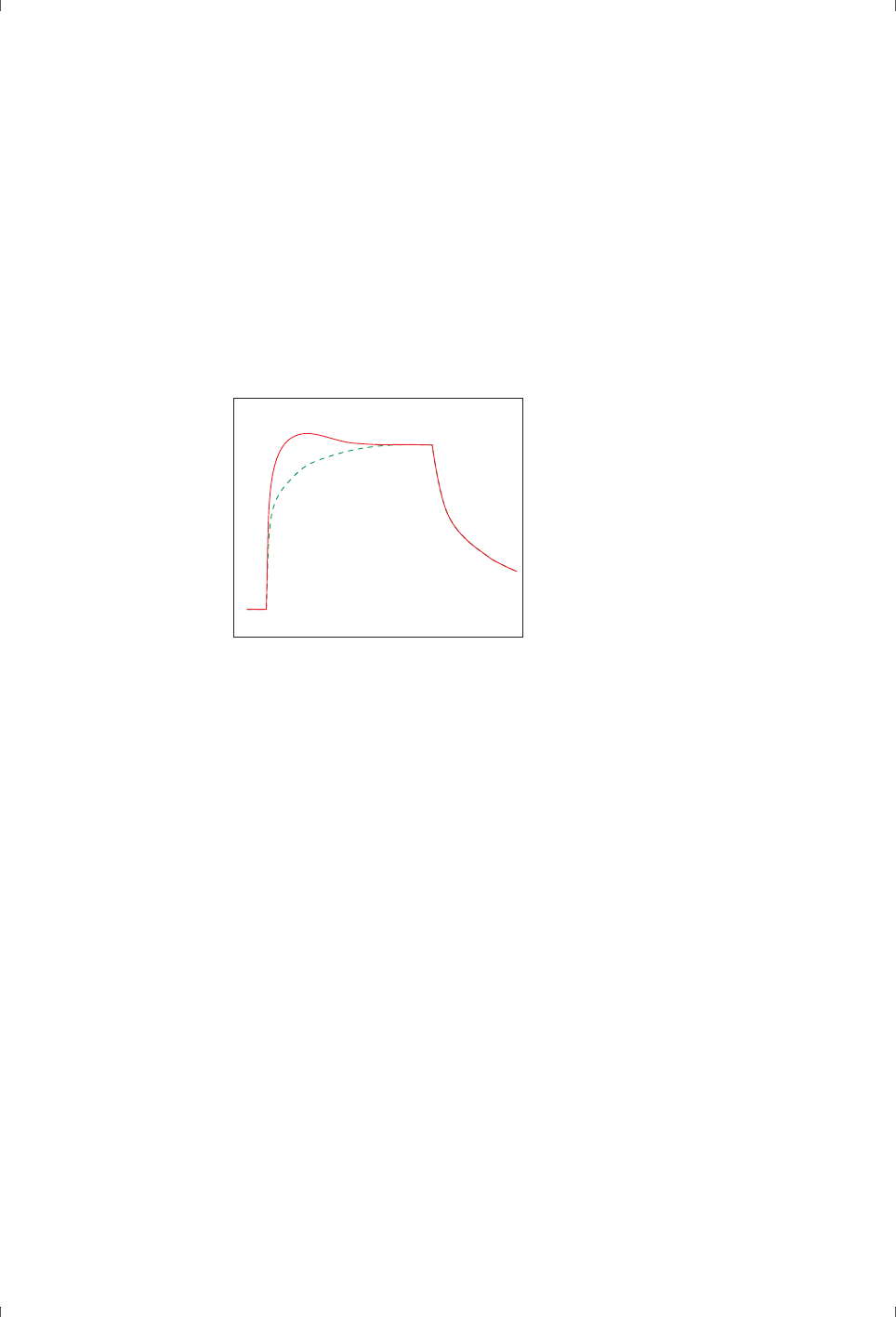
evidence of the interaction mechanism (Figure 2-1).
Figure 2-1. Fitting experimental data to a mathematical model does not prove that the
model is appropriate, and the same data may fit acceptably to several models. In this
example, the closeness of fit cannot distinguish confidently between a bivalent analyte
model (left) and a heterogeneous ligand model (right).

Biacore Assay Handbook 29-0194-00 Edition AA 11
Application overview 2
Assay set-up
There are currently two ways of setting up a kinetic analysis experiment (Figure
2-2):
• Multi-cycle kinetics runs each analyte concentration in a separate cycle,
regenerating the surface after each sample injection. It is important that
regeneration is optimized (see Biacore Sensor Surface Handbook for more
information) so that the surface properties are consistent from cycle to
cycle.
• Single-cycle kinetics runs a series of analyte concentrations in one cycle,
with no regeneration between sample injections. This approach is valuable
in situations where acceptable regeneration cannot be achieved, but is
more sensitive to response drift since the cycle time is longer.
Figure 2-2. In multi-cycle kinetics and affinity determinations (left), each sample is injected
in a separate cycle. The concentration series is presented as an overlay plot aligned at the
start of the injection in the evaluation software.
In single-cycle determinations (right), the samples are injected sequentially in the same
cycle.
Arrows in the illustrations mark the start of sample injections.
2.2.2 Affinity analysis
Interaction affinity may be determined in three independent ways using Biacore
systems: calculation from kinetic constants, measurement of steady-state
binding levels and determination of affinity in solution.
Affinity constants from kinetics
For simple 1:1 binding, the affinity constant is equal to the ratio of the rate
constants (equilibrium association constant K
A
= k
a
/k
d
, equilibrium dissociation
constant K
D
= k
d
/k
a
, see Appendix A). Values for affinity constants can therefore
be derived from kinetic measurements.
Note that these values, like the values for rate constants, are only valid in the
context of the model used to analyze the binding data. More complex binding
mechanisms do not always give straightforward relationships between affinity
and kinetics, and it may only be possible to derive affinity constants from kinetic
constants for values obtained with a simple interaction model.
2 Application overview
2.2 Kinetics and affinity measurements
12 Biacore Assay Handbook 29-0194-00 Edition AA
Steady state affinity
Analyzing the level of steady state or equilibrium binding as a function of
interactant concentrations is the foundation for many standard techniques for
determining interaction affinity. The basic relationship between concentrations
of interactants and complex and the affinity is described in Appendix A. Affinity
constants may be derived from the experimental data from linearized plots such
as Scatchard plots or (as supported in Biacore software) by direct analysis of a
plot of amount of complex against analyte concentration (Figure 2-3).
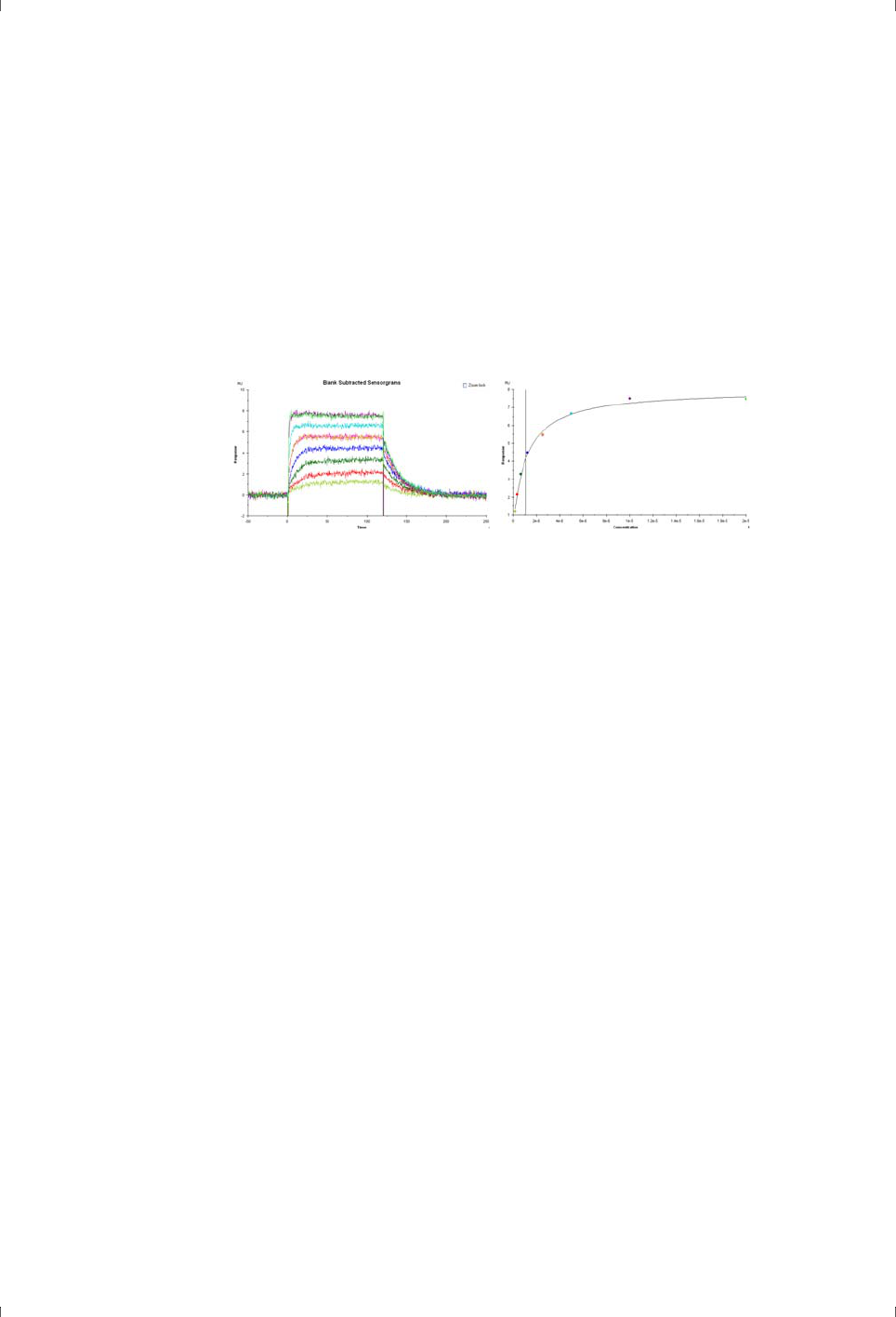
Figure 2-3. Sensorgrams (left) and corresponding plot of steady state response against
concentration (right) for determination of binding affinity. The vertical line in the right-
hand plot indicates the value of the calculated equilibrium dissociation constant K
D
.
Analysis of steady-state binding data by fitting the curve of binding level against
concentration can only be applied to 1:1 binding. A model for 1:1 binding at two
independent ligand sites can be useful in some circumstances, but more
complex models are not sufficiently robust for analysis of the data. It is however
possible to apply classical Scatchard analysis to data obtained from Biacore
systems, to give some indication of the causes of deviation from 1:1 binding
behavior.
It may be thought that the same experiment can in principle provide data for
both kinetics and steady state binding, offering two independent affinity values
from one set of data. In practice, however, restrictions on the injected volume of
sample (and therefore the contact time between sample and the sensor surface)
mean that interactions that give measurable kinetics seldom reach steady state
and vice versa. The affinity for a given interaction can therefore usually be
derived from kinetics or steady state measurements but seldom both.
Affinity in solution
The principle of measuring affinity in solution with Biacore systems is to
determine the free concentration (and therefore the concentration of complex)
in mixtures of known interactant concentrations, using the Biacore system for
the concentration assay. Commonly, the assay is set up with one interactant A
(or an analogue thereof) on the surface. A calibration curve is established for
determining the concentration of the second interactant B. Mixtures containing
variable concentrations of A and a fixed concentration of B are allowed to reach
equilibrium and then analyzed for the free concentration of B. The results are
analyzed using standard tools for affinity measurements. A requirement of this

Biacore Assay Handbook 29-0194-00 Edition AA 13
Application overview 2
approach is that only free B and not complex AB can bind to the surface-
attached A.
While the requirement for calibrated concentration measurements of analyte B
makes this approach somewhat cumbersome, the main advantage is that
samples can be incubated as long as necessary to reach steady state.
Measurements are therefore not limited by the available contact time between
sample and sensor surface.
2.3 Concentration measurements
Biacore systems can be used for measurement of analyte concentrations
because the amount of analyte that binds to the surface under appropriate
conditions can be related to the concentration of analyte in the sample. In
calibrated assays, the response is related to the analyte concentration through
a calibration curve, prepared with standards containing known analyte
concentrations. This approach is analogous to many other established methods
of concentration measurement. Calibration-free concentration analysis (CFCA) is
an alternative approach supported in some Biacore systems, where
concentrations are derived from the observed rate of diffusion-limited binding
without reference to a calibration curve.
Concentration assays using Biacore systems are described in detail in the
Biacore Concentration Analysis Handbook.
2.3.1 Calibrated assays
Calibrated concentration measurements may be designed as direct or indirect
assays.
• Direct assays measure analyte bound directly to the ligand on the sensor
surface. This approach is suitable for macromolecular analytes (molecular
weight > 5000 daltons): direct detection of smaller molecules is possible
but the useful range of the assay is generally limited in such cases.
Response enhancement or sandwich approaches can be used to amplify
the response obtained and/or to increase the selectivity of the assay.
• Indirect or competition assays provide an indirect measure of analyte
concentration, and are most useful for low molecular weight analytes. The
commonest format for indirect assays is the solution competition or
inhibition approach, where a known amount of an interacting partner
called the detecting molecule is mixed with the sample, and the amount of
free detecting molecule remaining in the mixture is measured. In the
surface competition method, analyte and a high molecular weight
analogue (often a protein conjugate) compete for binding to a common
partner on the sensor chip surface. In both competition assay formats, the
response obtained is inversely related to the concentration of analyte in
the sample.
2 Application overview
2.4 Mapping binding sites
14 Biacore Assay Handbook 29-0194-00 Edition AA
Measuring concentration in a calibrated assay, whether direct or indirect, relies
on the ability to establish a calibration curve using standards of known
concentration. The results for unknown samples are always obtained with
reference to the standard, and represent “true” concentrations only insofar as
the standard concentrations are reliable. More detailed discussion may be
found in the Biacore Concentration Analysis Handbook.
2.3.2 Calibration-free concentration analysis (CFCA)
Calibration-free assays are based on the relationship between the diffusion
properties of the analyte and the absolute analyte concentration. The
concentration is calculated from knowledge of the diffusion coefficient of the
analyte together with analysis of the observed binding rate under partially
diffusion-limited conditions. This approach can be useful in situations where no
satisfactory calibrant is available for the analyte under study, or when it is
important to determine “true” concentrations as closely as possible. Calibration-
free assays are always set up in the direct binding format.
The requirement for binding under partially diffusion-controlled conditions
limits the dynamic range of CFCA and restricts the approach to analysis of
macromolecules (molecular weight above 5,000 Da).
2.4 Mapping binding sites
Binding sites on a macromolecule can be mapped by testing the ability of
multiple binding partners to bind independently or to compete with each other.
Two binding partners that are directed towards distinct binding sites will be able
to bind simultaneously, while partners that are directed towards interfering
sites will compete with each other. This kind of analysis is most common in
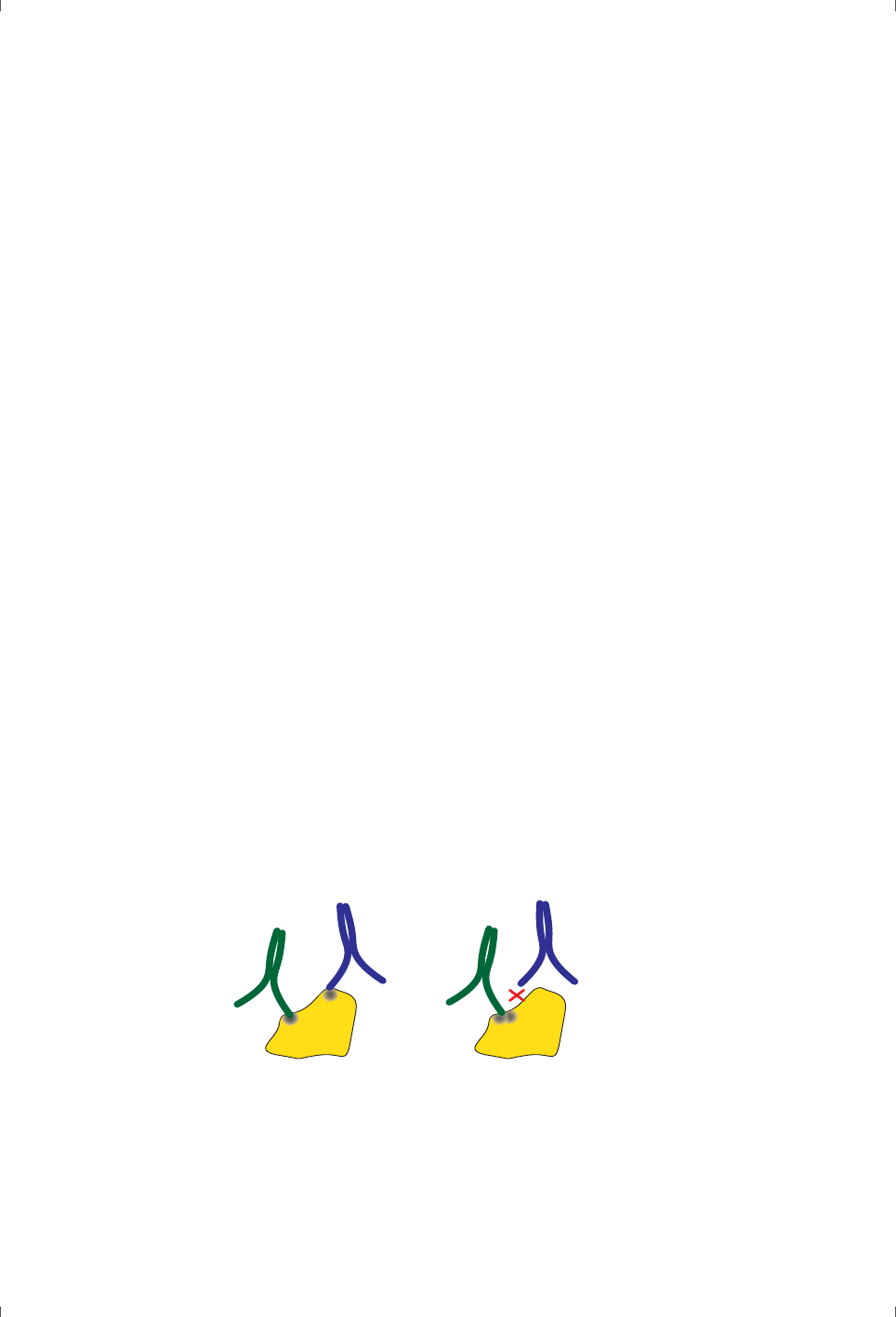
studies of antibody specificity (where it is called epitope mapping, see Figure
2-4), although it can in principle be applied to any investigation of multiple
binding sites on a macromolecule.
Figure 2-4. Different antibodies can bind simultaneously to distinct and spatially
separated epitopes (left), but not to common or overlapping epitopes (right).

Biacore Assay Handbook 29-0194-00 Edition AA 15
Application overview 2
Epitope mapping can be performed using two distinct approaches:
• Pair-wise binding tests the ability of pairs of antibodies to bind
simultaneously to the same antigen. Simultaneous binding is a clear
indication of distinct epitopes, while interference can result from binding
to the same or closely situated epitopes, or for example from a
conformational change in the antigen induced by binding of one antibody
that hides the epitope for the other.
• Peptide inhibition studies test the ability of peptides that represent specific
regions on the antigen to inhibit binding of single antibodies. A peptide
that blocks antibody binding is taken to represent at least a part of the
physical epitope for that antibody.

2 Application overview
2.4 Mapping binding sites
16 Biacore Assay Handbook 29-0194-00 Edition AA
Biacore Assay Handbook 29-0194-00 Edition AA 17
General considerations 3
3 General considerations
This chapter discusses aspects of working with Biacore systems that are
common to all application areas.
3.1 Sensor surfaces
3.1.1 General properties
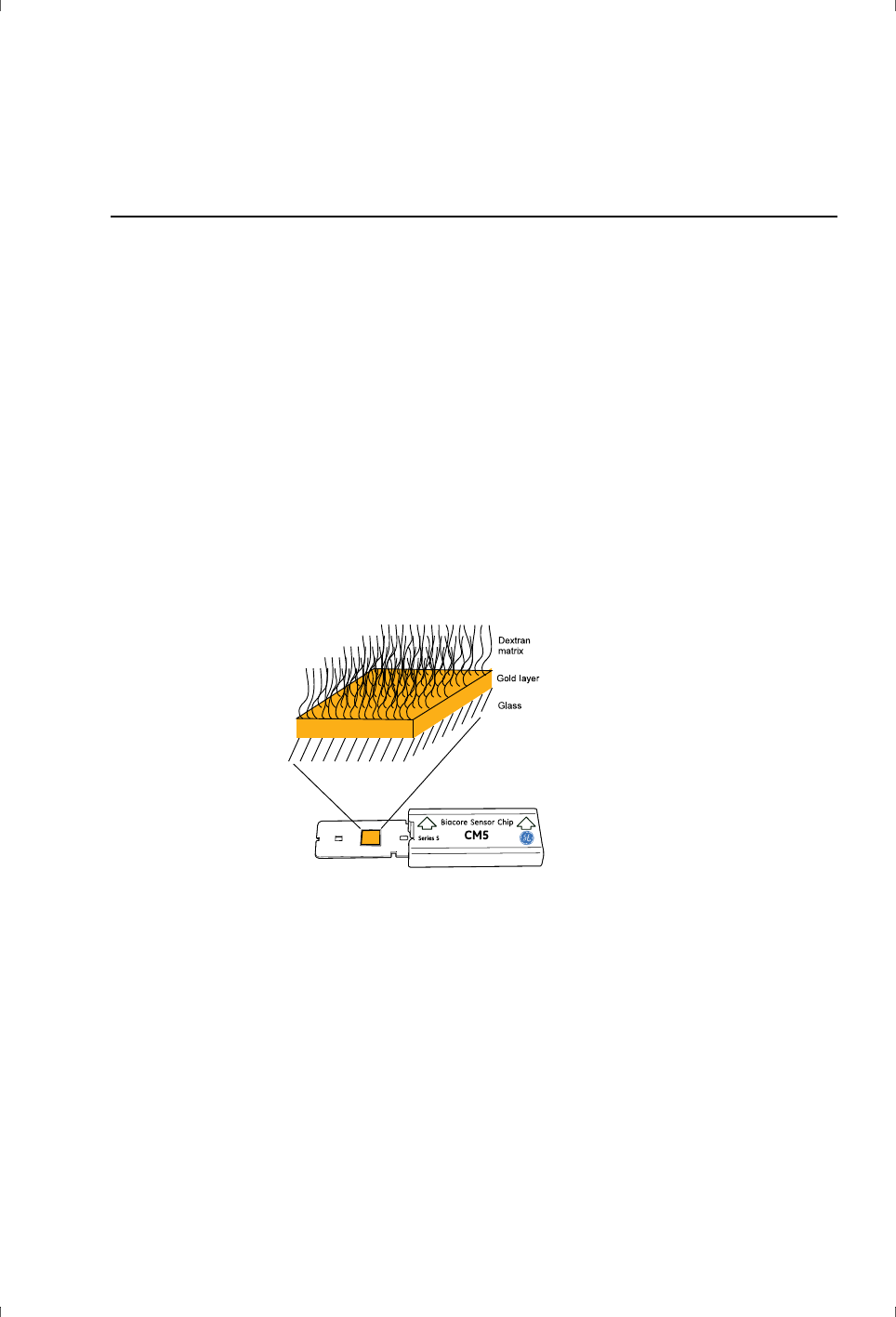
The sensor chip in Biacore systems consists of a glass slide coated with a thin
layer of gold. These components, together with the docking system for
mounting the sensor chip in the optical system, are required for generation of
an SPR signal. To provide a suitable environment for the molecular interactions
being studied, the gold surface is covered by a linker layer and (on most sensor
chip types) a matrix of modified dextran (Figure 3-1). It is the surface matrix that
determines the properties of the sensor chip with respect to ligand attachment
and molecular interaction.
Figure 3-1. Schematic illustration of the structure of the sensor chip surface on CM-series
chips.
3.1.2 Sensor surface types
GE Healthcare offers a range of sensor surfaces designed for different
application requirements, as summarized in Table 3-1. Detailed descriptions of
sensor chip types may be found in the Biacore Sensor Surface Handbook and
on www.gelifesciences.com/biacore.
3 General considerations
3.1 Sensor surfaces
18 Biacore Assay Handbook 29-0194-00 Edition AA
Table 3-1. Characteristics and application areas for sensor chips.
Sensor Chip Characteristics Use
CM-series sensor chips (carboxymethyl-dextran surface matrix)
CM5 Moderate capacity General purpose
CM7 High capacity High immobilization levels for
work with low molecular
weight analytes (not suitable
for protein analytes or capture
applications).
CM4 Low capacity
(low substitution density)
Low immobilization levels for
kinetics.
May help to reduce non-
specific binding from complex
samples.
CM3 Low capacity
(shorter dextran chains)
Large particulate analytes
(viruses and cells).
Low immobilization levels for
kinetics.
May help to reduce non-
specific binding from complex
samples.
C1 Flat surface, no dextran
matrix. Carboxyl groups
directly attached to the
surface linker layer.
Restricted mobility of
attached ligand.
Large particulate analytes
(viruses and cells).
Hydrophobic surfaces
HPA No surface matrix or
attached groups
Direct adsorption of lipid
monolayers, with or without
associated proteins.
L1 Dextran matrix
substituted with
lipophilic residues
Capture of intact lipid vesicles.

Biacore Assay Handbook 29-0194-00 Edition AA 19
General considerations 3
3.2 Attaching the ligand
In broad terms, there are two main approaches to attaching macromolecular
ligands to the sensor surface, covalent immobilization and high affinity capture.
Details of these alternatives may be found in the Biacore Sensor Surface
Handbook.
3.2.1 Covalent immobilization
Covalent immobilization involves irreversible chemical attachment of the ligand
to the surface, usually to the carboxymethyl groups on CM-series sensor chips.
The ligand remains on the surface throughout the lifetime of the sensor chip,
and is regenerated to remove non-covalently bound molecules after each
analysis cycle.
Covalent immobilization may exploit amine, thiol (either native or introduced) or
aldehyde groups (obtained by oxidation of cis-diols) on the ligand. Amine
coupling is the most widely used alternative.
Conditions for immobilization
The concentration of ligand in the surface matrix for analysis in Biacore systems
is many times higher than that in a typical bulk solution: immobilization of a
protein at a response level of 100 RU on Sensor Chip CM5 corresponds roughly
to a bulk concentration of 1 mg/ml in the surface matrix, while typical bulk
concentrations used for immobilization are 50-100 µg/ml. In most
immobilization methods, ligand is concentrated on the surface by a process of
electrostatic pre-concentration. This occurs when the pH of the ligand solution is
below the isoelectric point of the ligand, so that the ligand carries a net positive
charge, but above the pKa (3.5) of carboxyl groups on the surface so that the
Surfaces for ligand capture
SA Streptavidin covalently
attached to dextran
matrix
Permanent capture of
biotinylated molecules
CAP Oligonucleotide
covalently attached to
dextran matrix
Reversible capture of
biotinylated ligands, mediated
by a streptavidin-carrying
complementary
oligonucleotide.
NTA Nitrilotriacetic acid
covalently attached to
dextran matrix
Capture of histidine-tagged
ligands
Sensor Chip Characteristics Use
3 General considerations
3.2 Attaching the ligand
20 Biacore Assay Handbook 29-0194-00 Edition AA
surface is negatively charged. Low ionic strength favors the electrostatic
interaction, and buffers with 10-20 mM total cation concentration are generally
recommended.
Choice of immobilization chemistry
Many macromolecules can be immobilized as ligands on the sensor surface
without significantly interfering with the interaction being studied, provided that
the coupling chemistry involves groups that are distant from the site of
interaction. Chemistries such as amine coupling that target common groups
may result in a mixture of ligand molecules on the surface with different points
of attachment: in some cases, ligand may be partially inactivated with respect
to interaction by the attachment, but this does not matter as long as sufficient
active molecules are present. More specific chemistry such as thiol coupling
may be used to reduce the variation in orientation of the attached molecules, or
even to determine precisely how the molecules will be attached (if the ligand
contains only a single target group for attachment).
Figure 3-2. Amine coupling is frequently successful, but in this example (immobilization of
a hormone receptor) amine coupling destroys the ligand activity completely. The best
results are obtained with high affinity capture.
Amine coupling
Immobilized: 10,000 RU
Activity: undetectable
Thiol coupling
Immobilized: 12,000 RU
Activity: 5%
Poly-histidine tag capture
Immobilized: 2,000 RU
Activity: 20%

Biacore Assay Handbook 29-0194-00 Edition AA 21
General considerations 3
Amine coupling is generally applicable to a wide range of protein ligands, and is
often the most convenient method of attachment. However, testing a range of
attachment chemistries can be worthwhile in critical cases: Figure 3-2
illustrates results from immobilization of a hormone receptor where amine
coupling gave a fully acceptable immobilization level but activity was preserved
better by thiol coupling. High affinity capture of poly-histidine tagged receptor
on Sensor Chip NTA gave however the best yield in terms of binding activity
(tested by binding of hormone) per immobilized RU.
3.2.2 High affinity capture
High affinity capture refers to attachment of ligand by binding to an
immobilized capturing molecule, and offers advantages over covalent
immobilization in a number of situations:
• Capture does not require any chemical modification of the ligand and can
be performed in physiological buffer conditions. It can therefore be used
with ligands that are not amenable to covalent immobilization.
• Normally, regeneration of the surface with a captured ligand involves
removal of ligand together with other bound molecules, leaving the
capturing molecule ready bind fresh ligand for the next cycle. While this
procedure increases consumption of ligand, it removes the requirement
that ligand is not damaged by regeneration and allows the ligand to be
changed between analysis cycles on the same sensor chip.
• Capture may be used to attach a specific ligand from a complex sample,
provided the capturing interaction is sufficiently selective. Covalent
immobilization of ligand from such samples would require prior
purification of the ligand.
Conditions for ligand capture are dictated by the requirements of the capturing
interaction, and capturing is normally performed in the same buffer conditions
as the interaction being studied. It is however important that the capturing
interaction is reasonably stable under the chosen conditions, so that ligand
does not dissociate significantly during sample analysis. Demands on capturing
stability vary with different applications, and are highest for kinetic and affinity
analyses, since loss of ligand will cause a fall in response as well as reducing the
analyte binding capacity of the surface. Simpler applications such as detection
and screening may tolerate more loss of ligand during sample injection.
3.2.3 Response levels
One of the issues in preparing the sensor surface for a given application is the
question of how much ligand that should be attached. All steps in the
attachment process are monitored in the instrument, and the amount of
attached ligand is in principle given directly by the response above baseline
(Figure 3-3).
3 General considerations
3.2 Attaching the ligand
22 Biacore Assay Handbook 29-0194-00 Edition AA
Figure 3-3. Sensorgram from a typical amine coupling, illustrating the distinction between
the amount of ligand bound and the amount immobilized.
Note: Even activation of the surface with EDC/NHS can be detected as a small
increase in response. When the response from immobilized ligand is low,
it may be more appropriate to estimate the ligand level from the response
after activation rather than the baseline at the start of the immobilization
procedure.
A more relevant parameter in designing and preparing a surface is however the
analyte binding capacity (called
Rmax
, for maximum response), since this will
determine the expected response range from samples. The amount of ligand on
the surface as measured from the increase in response gives an indication of
the theoretical analyte binding capacity of the surface, provided that the relative
sizes of ligand and analyte are known. Assuming a 1:1 binding stoichiometry:
The practical analyte binding capacity is obtained experimentally by injecting
analyte at a high concentration to saturate the surface (or by extrapolating
binding levels from measurements at a series of concentrations). Comparison of
the theoretical and practical R
max
values provides a measure of the activity of
surface-attached ligand. Protein preparations are seldom 100% active, and for
generic attachment methods such as amine coupling, activities in the region of
70% and above may be expected.
The shape of the sensorgram from the attachment procedure can provide
guidance in choosing appropriate conditions. Bear in mind that the amount of
ligand immobilized will generally be less than the amount bound during
preconcentration. Most Biacore systems support a mode of ligand
immobilization using amine coupling that exploits the information from a brief
R
max
RU
analyte MW
ligand MW
----------------------------- -
immobilized ligand level (RU)=

Biacore Assay Handbook 29-0194-00 Edition AA 23
General considerations 3
preconcentration injection to adapt the immobilization process to reach a
specified level.
• The sensorgram slope and shape during protein injection indicates the
efficiency of electrostatic preconcentration. Try to use a combination of
ligand concentration and injection time so that the amount bound reaches
a plateau. This will contribute to the robustness of the immobilization
procedure.
• The relative response levels before and after deactivation of the surface
indicate how well the electrostatically bound protein has become
covalently attached. Deactivation washes out non-covalently bound
material: a reduction in response may be observed after deactivation but
this does not affect the assay as long as sufficient ligand is attached to the
surface.
If the immobilization works in principle satisfactorily but the amount of ligand
attached is not suitable for the assay, adjust one or more of the parameters
ligand concentration, immobilization pH, ligand injection time and surface
activation time.
3.3 General buffer considerations
3.3.1 Buffer substances
In general, Biacore systems are compatible with most buffer substances used in
biological studies. Exceptions may be dictated by specific properties of the
interacting molecules or by the situation where the buffer is used (for example,
the primary amine groups in Tris preclude the use of this buffer for attaching
ligand by amine coupling).
HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pK
a
7.55) is a
satisfactory buffer substance for many protein-protein interactions, and ready-
to-use buffers based on 10 mM HEPES are available for use with Biacore
systems.
Phosphate-based buffers are often more suitable for work with low molecular
weight analytes since organic buffers such as HEPES can bind to the ligand and
interfere with accurate detection of low molecular weight compounds.
3.3.2 Ionic strength
Physiological ionic strength buffers (containing 0.15 M monovalent cations) are
recommended unless other requirements are imposed by the interaction being
studied. Reducing the ionic strength will increase the tendency for non-specific
electrostatic binding of molecules from the sample to the sensor surface. Higher
ionic strength may be tested if non-specific binding is a problem.
3 General considerations
3.3 General buffer considerations
24 Biacore Assay Handbook 29-0194-00 Edition AA
3.3.3 Additives
Detergent
Inclusion of non-ionic detergent (0.05% Surfactant P20, Tween™ or equivalent)
in all buffers is recommended to minimize deposition of protein and other
biomolecules in the flow system. The detergent should be included at a
concentration above the critical micelle concentration (CMC). At lower
concentrations, the detergent itself can accumulate in the flow system and lead
to unwanted response effects as it is released (see FIgure 3-4).
Figure 3-4. Schematic illustration of typical “detergent effects” caused by using detergent
at concentrations below CMC. The broken line shows an undisturbed sensorgram for
comparison.
Some applications, particularly those involving lipid vesicles and/or
hydrophobic proteins, require detergent-free buffer. In such cases it is essential
to clean the flow system regularly and carefully using the appropriate
maintenance tools provided with the system, in order to ensure continued high
performance. Poor instrument cleaning can result in severely disturbed
sensorgrams that are difficult to interpret. A severe example is shown in Figure
3-5.
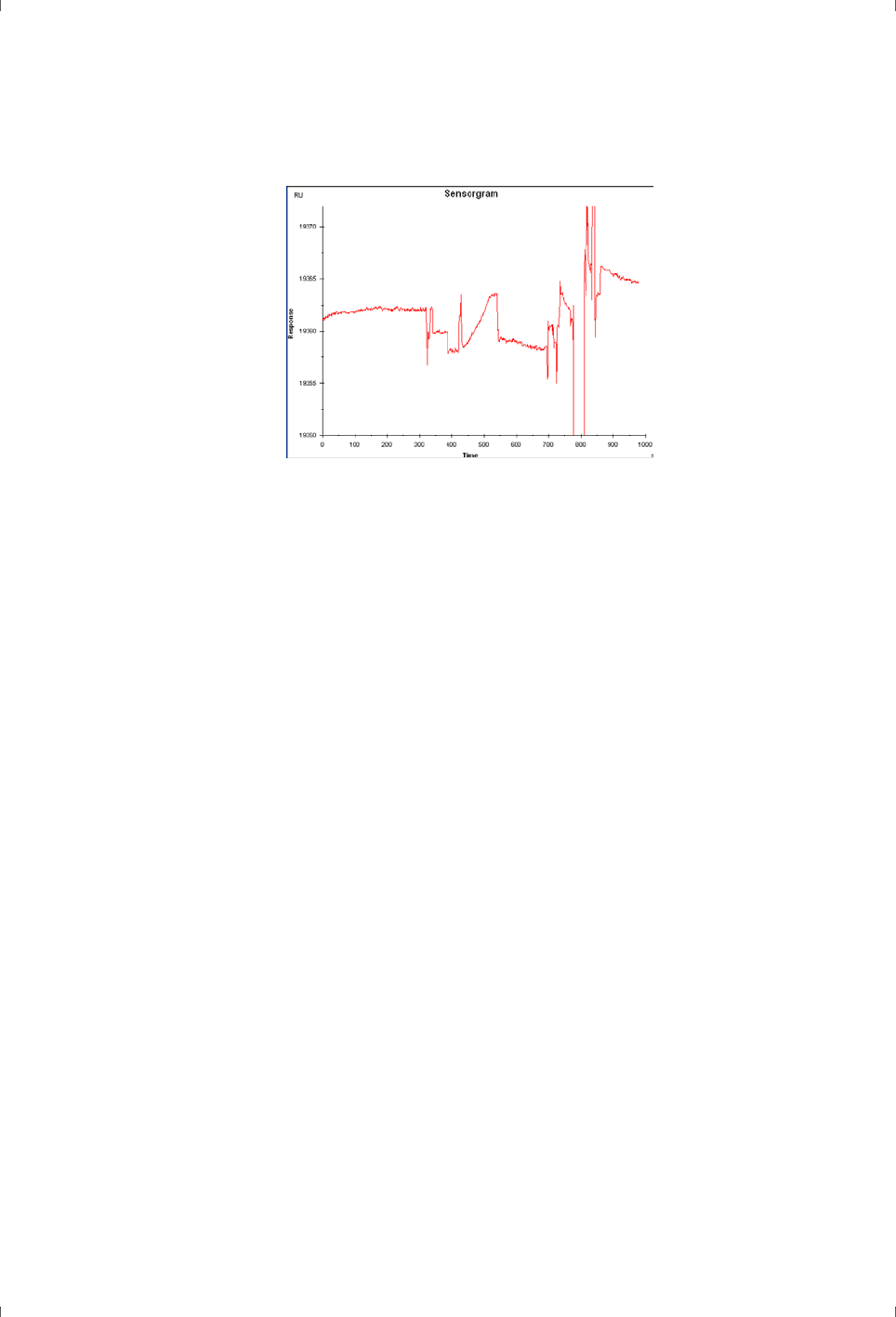
Biacore Assay Handbook 29-0194-00 Edition AA 25
General considerations 3
Figure 3-5. Severe example of the consequences of poor instrument cleaning.
EDTA
HBS-EP and HBS-EP+ general purpose buffers from GE Healthcare include 3 mM
EDTA to remove traces of free divalent metal ions that may be present in the
buffer. This is a general precautionary measure and is not specifically required
for work with Biacore systems. Buffers with no added EDTA are also available.
NSB Reducer
Addition of NSB reducer (a preparation of soluble carboxymethyl dextran) to
samples can in some cases help to reduce non-specific binding of sample
components to the dextran matrix on the sensor surface. The recommended
concentration of NSB Reducer is 1 mg/ml. NSB Reducer should only be used in
situations where non-specific binding is a problem (typically with complex
samples such as serum or whole cell extracts).
Organic solvents
Many low molecular weight organic compounds, relevant particularly in
pharmaceutical development work, are sparingly soluble in aqueous buffers
and require addition of organic solvents. Dimethyl sulfoxide (DMSO) is commonly
used. Biacore systems can be used with buffers containing up to 10% DMSO.
However, organic solvents contribute significantly to the bulk refractive index of
samples and buffers: 1% DMSO contributes approximately 1200 RU to the
response level. Expected responses from low molecular weight analytes, which
often need organic solvents to maintain solubility, may be as low as 10-20 RU or
less. It is therefore crucial that the measured responses are accurately
compensated for any variations in organic solvent concentration. Procedures
for adjusting measured sample responses for solvent effects, called solvent
correction, are described in principle in Appendix B and in more detail in the
documentation for Biacore systems where the correction is supported.
3 General considerations
3.3 General buffer considerations
26 Biacore Assay Handbook 29-0194-00 Edition AA
Other additives
Avoid additives that can compete with the ligand for immobilization chemistry,
for example sodium azide with amine coupling chemistry. Even at low
concentrations, sodium azide can effectively prevent amine coupling of
proteins.
3.3.4 Buffer preparation
All buffers regardless of composition should be filtered before use and degassed
if the instrument does not have a built-in degasser. Disturbances caused by
particles and microscopic air bubbles introduced over the sensor surface
appear typically as spikes in the sensorgram. Air bubbles or particles that
remain on the surface can result in more long-lived displacements in the
response (Figure 3-6).
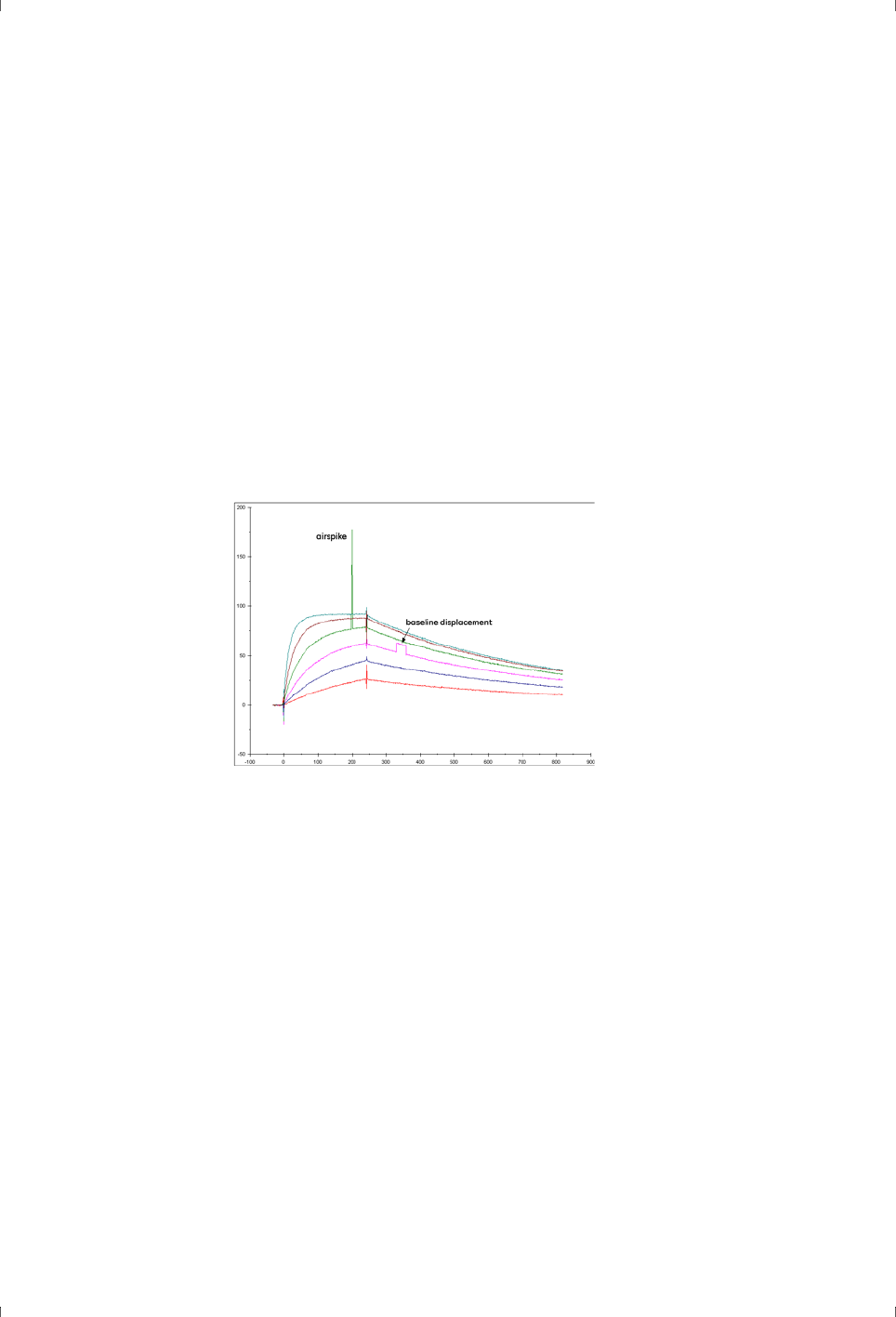
Figure 3-6. Airspikes and transient baseline displacements are often attributable to
inadequately degassed running buffer.
Filtering
Buffers should be filtered through a 0.22 µm filter to remove particles.
Serum and plasma samples should be centrifuged or filtered to remove
aggregated material, particularly if the samples have been frozen.
Degassing
Degas running buffer before use unless you are working with a Biacore system
fitted with an in-line degasser. Alternatively use degassed ready-to-use buffers
from GE Healthcare.
Always degas buffer solutions before adding detergent to avoid frothing.
Biacore Assay Handbook 29-0194-00 Edition AA 27
General considerations 3
3.4 Matching sample and running buffer
3.4.1 Matching refractive index
Differences in refractive index between sample and running buffer give rise to
so-called bulk shifts in sensorgrams at the beginning and end of sample
injection. The bulk shift may be negative or positive depending on the difference
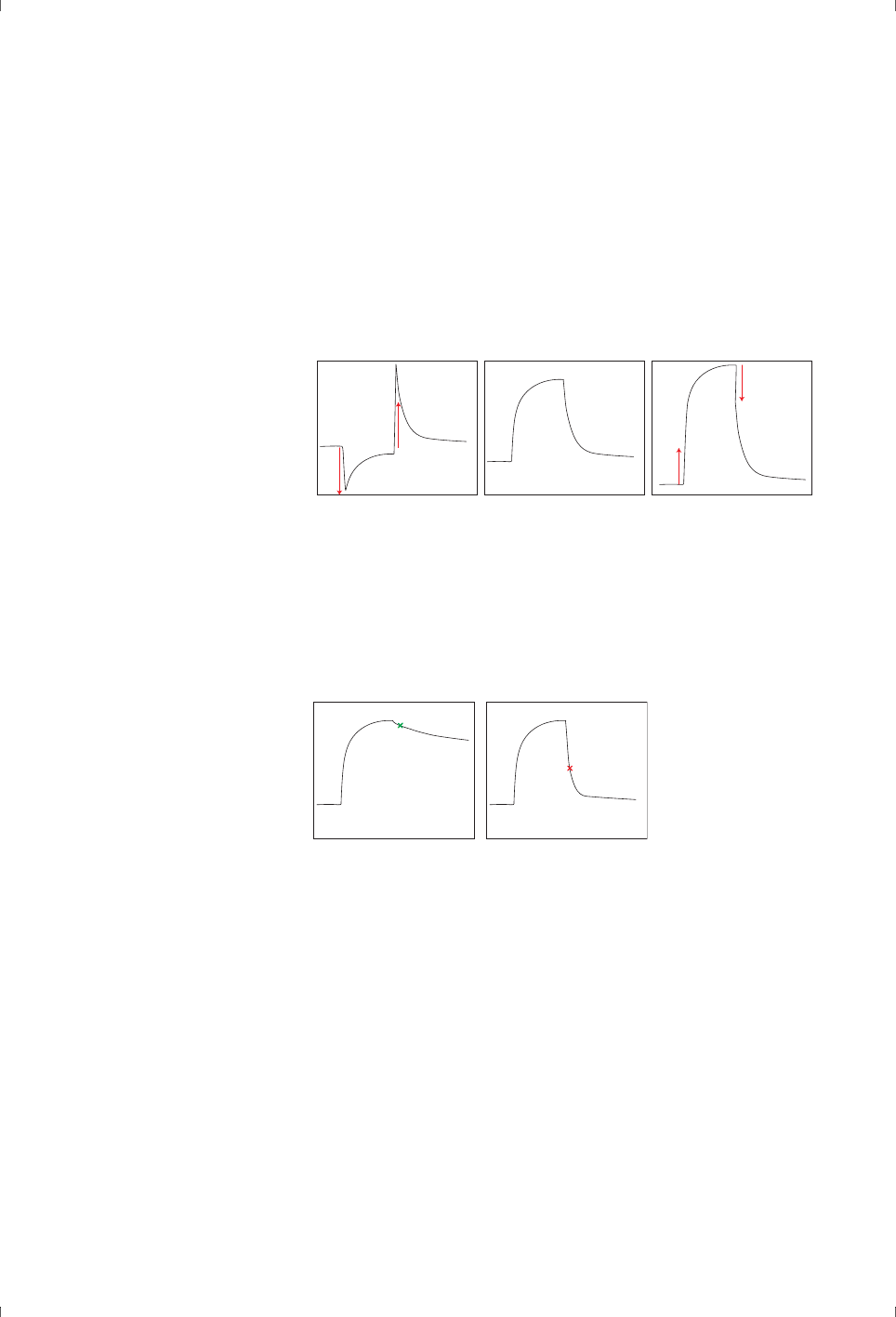
in refractive index (Figure 3-7).
Figure 3-7. Schematic illustration of bulk shifts: negative (left), zero (center) and positive
(right).
Some applications, such as concentration measurement and in some cases
screening, rely on single-point measurements (report points) that can be placed
after the end of the sample injection. These measurements are not affected by
bulk shifts so that accurate matching sample and running buffer is not
necessary (Figure 3-8).
Figure 3-8. If dissociation is slow enough, binding levels can be assessed from a report
point placed shortly after the end of the sample injection (left). However, if dissociation is
rapid (right) a report point at this position will not give a reliable measure of the binding
level.
In contrast, applications that rely on response measurement during sample
injection need to deal with the question of bulk response. Careful matching of
sample and running buffer with respect to refractive index goes a long way to
eliminating bulk shifts, but exact matching is seldom practicable. Subtracting
the response from a reference surface usually handles bulk shifts satisfactorily
provided that the shifts are reasonably small in relation to the measured analyte
responses. Special measures, called solvent correction, are however required for
applications where bulk shifts can be large and expected responses are small
(typically assays involving low molecular weight organic analytes, such as
response
time
response
time

3 General considerations
3.5 Dilution series and replicates
28 Biacore Assay Handbook 29-0194-00 Edition AA
screening for potential drug development candidates). These special
considerations are dealt with in Appendix B.
It is not always possible to match the refractive index of sample and running
buffer, for example in analysis of unpurified samples such as serum. In such
cases it is important that measurements are made after the end of the sample
injection to avoid bulk shift effects.
3.4.2 Matching buffer composition
Matching of sample and running buffer composition is particularly important for
kinetic measurements, regardless of the question of bulk response shifts, since
the sensorgram contains information about dissociation kinetics both during
and after sample injection. For correct analysis of the kinetics, it is important
that the dissociation process during and after sample injection occur in the
same environment.
3.4.3 Matching buffer environment in practice
Several approaches can be used to match sample and running buffer,
depending on the form in which the original sample is available.
• Samples that are available in sufficiently concentrated stock solution may
be diluted in running buffer so that the concentration of other components
in the stock is negligible. The minimum dilution factor needed to achieve
this depends on the composition of the stock solution and the stringency
of the buffer matching requirements for the assay at hand.
• Buffer exchange techniques such as desalting columns or dialysis are
often the most effective way to prepare samples in running buffer. Buffer
exchange can be essentially complete with minimum dilution of the
sample, and commercially available solutions such as micro-spin columns
allow rapid preparation of small sample volumes.
• Solid samples may be dissolved directly in running buffer: filter or
centrifuge the solution if there is a risk that undissolved material remains.
Be aware however that many commercially available proteins supplied in
solid form include considerable amounts of salt or other stabilizing
medium. Dissolving a dry preparation in running buffer does not
guarantee that the composition or refractive index of sample and buffer
will be matched.
3.5 Dilution series and replicates
Recommendations in this section relate to factors that should be standard in
laboratory work. They are however included here as a reminder, since they can
in some instances be critical to the success of the application.

Biacore Assay Handbook 29-0194-00 Edition AA 29
General considerations 3
Dilution series play an important role in a number of contexts in work with
Biacore systems: for example, kinetics and affinity determinations require
analysis over a range of analyte concentrations, and calibrated concentration
assays use dilution series for establishing a standard curve and also for
investigating certain performance characteristics such as parallelism. In many
situations, performing replicate determinations to establish reproducibility is a
given component in assay design. The way in which dilution series and
replicates are prepared and analyzed can have a significant impact on the value
of the results.
3.5.1 Dilution series
There are various possible sources of error in creating a dilution series, and the
best approach depends on the available equipment and the volumes involved
as well as the accuracy demands of the application.
Reliable pipettes are essential for creating accurate dilution series by volume.
Where possible (particularly for small volumes), use repeated aliquots of the
same volume for dilution series, to avoid errors that may be introduced by
repeatedly changing the volume on variable volume pipettes.
Two different basic methods may be used:
• Serial dilution where each dilution is used as the sample solution for the
next step.
• Parallel dilution where sample is always taken from the same stock
solution and mixed with increasing volumes of buffer.
Using the same fixed volume pipettes for both sample and buffer provides the
best accuracy for dilution factor (although not necessarily for absolute
volumes). If different pipettes are used with different errors of delivery, the
dilution error will propagate through the series more significantly with serial
dilution than with parallel. However, serial dilution is a simpler procedure,
requiring fewer operations with a fixed-volume pipette, so that parallel dilution
is more susceptible to errors of reproducibility.
For the best accuracy in preparing dilution series, volumes of buffer and sample
used may be determined by weighing and the dilution factor calculated
individually for each sample. This avoids issues of both calibration accuracy and
reproducibility for pipetted volumes.
3.5.2 Replicates
Measuring replicate samples to establish the reproduciibility of an assay should
be a standard procedure in all laboratory work, but it is not always clear how the
replicates should be prepared and measured in order to provide the most useful
information. Replication can cover the whole assay procedure from raw sample
material to evaluation of the final results, or it can focus on one or more specif ic

3 General considerations
3.6 Preparing vials and microplates
30 Biacore Assay Handbook 29-0194-00 Edition AA
components of the procedure. Different assay situations will require different
approaches in this respect.
Replicate samples designed to test the reproducibility of a Biacore-based assay
should be prepared and measured according to the following general
guidelines:
• Take replicates from a well-defined step in the sample preparation
procedure, with a clear awareness of what parameters are being tested
for reproducibility. For example, taking replicate samples from the same
tube in a dilution series will test the reproducibilty of the assay procedure
itself, whereas preparing replicate dilution series from a common stock
solution will include pipetting and dilution procedures in the repliacte test.
• Place replicate samples in separate vials or microplate wells for the assay
wherever possible, so that only one sample is taken from each position.
Injecting multiple samples from the same autosampler position is not
recommended since evaporation from vials or wells that have been
penetrated can affect the concentration.
• Analyze replicates at well-spaced intervals in the assay, to provide a check
on any drift in the assay performance with time. In many assays, this
factor can be conveniently tested by using repeated measurements on
control samples: in some cases however (such as kinetics and affinity
analysis) it can be valuable to apply the principle to replicate
concentrations of the sample.
3.6 Preparing vials and microplates
Samples and reagents are loaded into sample racks or microplates (according
to the requirements of the particular Biacore instrument). All samples for one
assay run are normally loaded at the start of the run: if samples have a tendency
to precipitate with time, bear in mind that the samples that are analyzed last
may have been standing for several hours in the autosampler.
Beware of using samples that might precipitate or aggregate with time (for
example, low molecular weight organic compounds close to the limit of
solubility).
Cover samples as soon as they are prepared, and keep them covered for the
duration of the assay to prevent evaporation. Always use vial caps and
microplate foil for Biacore systems from GE Healthcare: the autosampler needle
may not penetrate caps or foil from other sources satisfactorily, resulting in
failed assays or even damage to needle. When sealing microplates with foil,
make sure the foil is correctly placed so that the wells are covered by areas free
from adhesive. If the foil is wrongly applied, adhesive may accumulate on the
needle and cause malfunction.
Biacore Assay Handbook 29-0194-00 Edition AA 31
Screening and detecting binding partners 4
4 Screening and detecting binding partners
Screening applications using Biacore systems fall into two main categories that
differ significantly in challenges and experimental design:
• Small molecule (LMW) screening, where response levels are low and
organic solvents are often needed to maintain analyte solubility. These
interactions are usually rapid and regeneration is not needed.
• Biopharmaceutical (frequently antibody) screening where response levels
are higher but the sample matrices (cell culture supernatant or cell
extracts) are often complex. These interactions are usually slower:
regeneration is simplified if a capture approach is used.
Table 4-1 summarizes the main experimental conditions for LMW and antibody
screening.
Table 4-1. Main experimental conditions for LMW and antibody screening.
1
Recommended 2% unless higher concentration is required to maintain substance
solubility.
A third related application area is immunogenicity testing, which involves
detecting anti-drug antibodies (ADAs) in plasma or serum samples. Here the
simultaneous presence of ADAs and drug in the samples creates special
requirements.
LMW screening Antibody screening
Buffer Phosphate HEPES
Additives:
- DMSO
- detergent
Yes (1 to 10%)
1
Yes
No
Yes
Flow rate Typically 10 µl/min Typically 10 µl/min
Contact time 15 to 30 s 1 to 2 min
Report point Before end of sample
injection
After end of sample
injection
Regeneration Usually not needed Usually needed

4 Screening and detecting binding partners
4.1 Small molecule screening
32 Biacore Assay Handbook 29-0194-00 Edition AA
4.1 Small molecule screening
4.1.1 Goals
The primary goal of small molecule screening in pharmaceutical development
is to identify candidate molecular structures for further development, on the
basis of their binding to selected target molecules. The first stages of the
process is often screening of candidate libraries or fragment libraries to identify
promising binders for further development and refinement. Information-rich
screening with Biacore systems can provide a wealth of valuable data in
addition to yes/no binding results, including comparative binding to multiple
targets to identify promiscuous (and therefore less interesting) binding
candidates and assessment of kinetics and affinity properties that become
increasingly important as the candidate molecules are refined.
4.1.2 Sensor surface preparation
The response obtained from LMW analytes is inherently low because of the
molecular size: in addition, binding affinities are often weak (particularly for
fragment screening) which further reduces the expected response levels. For
this reason, sensor surfaces for LMW screening are prepared with high levels of
ligand (typically 8,000 to 10,000 RU for average-sized proteins).
Accurately determining the activity of the immobilized ligand requires high
analyte concentrations to saturate the surface with weak binders (typically 0.1
to 2 mM for fragments and 0.05 to 0.2 mM for LMW compounds). Testing binding
activity with a positive control is desirable if a suitable control analyte is
available.
4.1.3 Sample preparation
In the first phases of small molecule screening, samples are often prepared at a
fixed concentration in the same buffer. There is usually no opportunity to
optimize concentration or buffer conditions.
Many small organic compounds are sparingly soluble in aqueous buffers and
require inclusion of organic solvents (commonly 1% to 3% DMSO) to maintain
solubility. DMSO has a strong refractive index contribution (about 1200 RU for
1% DMSO), and both careful matching of sample and running buffer and
correction for small differences in DMSO concentration are required (see below).
4.1.4 Buffers
Phosphate buffers are generally recommended for work with small molecules.
Using organic buffers such as HEPES can bind to the ligand and interfere with
accurate detection of small organic compounds. Physiological ionic strength
(150 mM monovalent cations) should be used to reduce non-specific binding of

Biacore Assay Handbook 29-0194-00 Edition AA 33
Screening and detecting binding partners 4
compounds to the sensor surface, and inclusion of detergent (0.05% Surfactant
P20) generally improves data quality by reducing drift and signal disturbances.
The composition of running buffer and sample buffer must be matched as
closely as possible, particularly with respect to organic solvent concentration.
Variations in DMSO content between samples resulting from both evaporation
and absorption of water are however unavoidable. Procedures that correct for
differences in bulk refractive index between samples, called solvent correction,
are described in Appendix B and are implemented in Biacore systems designed
for work with small molecules.
4.1.5 Analysis conditions
Flow rate
Use low flow rates (10 µl/min) for LMW screening applications to conserve
sample. Issues such as mass transfer limitations (Section A.1) and resolution of
fast binding events are not relevant.
Contact time
Most interactions involving small molecules are rapid, and contact times can be
kept short (30 to 60 seconds) to increase screening throughput.
Regeneration
Regeneration is generally not necessary in LMW screening applications, since
dissociation is rapid and is usually complete within 1 minute or thereabouts.
Report points
Because of the rapid dissociation of many LMW compounds from the target
molecule, report points for screening should be placed before the end of the
sample injection. LMW compounds sometimes show rapid binding to specific
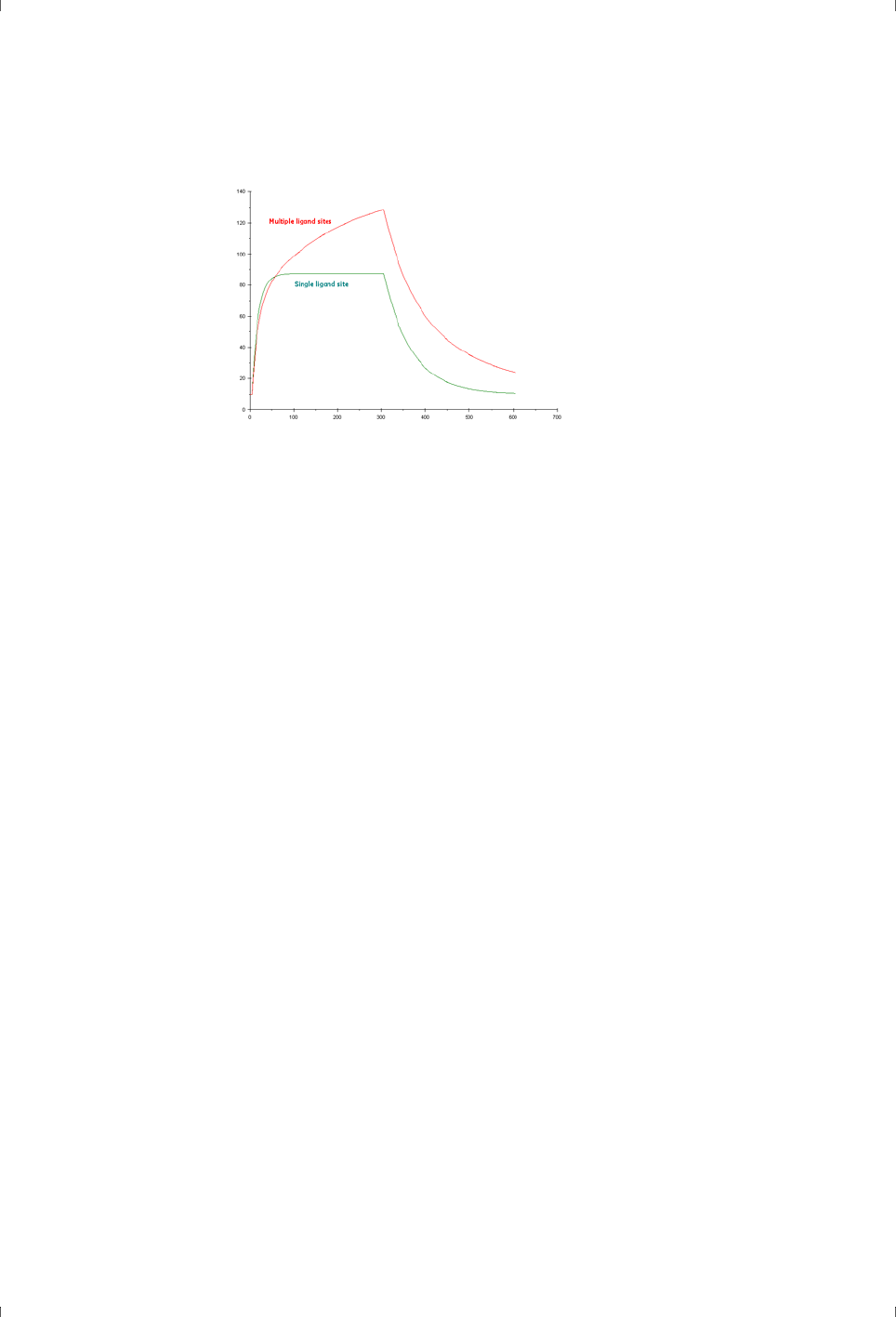
sites followed by slower and more indiscriminate binding (Figure 4-1). Placing
report points early in the sample injection can reduce the impact of this
behavior and give a more accurate indication of specific binding ability.
4 Screening and detecting binding partners
4.1 Small molecule screening
34 Biacore Assay Handbook 29-0194-00 Edition AA
Figure 4-1. Simulated sensorgrams illustrating 1:1 binding (dark green) and binding to one
specific site followed by slower and more indiscriminate binding (red).
Other considerations
Some LMW compounds are “sticky” and can be difficult to wash off the sensor
surface and out of the flow system, causing carry-over of material to the next
analysis cycle. This can be detected by routinely including a “carry-over
injection” of buffer after the sample injection: the response from a “sticky”
compound will be carried over into this buffer injection. Include a flow system
wash with 50% DMSO after every cycle to minimize problems with sticky
compounds. If a screening experiment involves compounds that are known to
be sticky, place these at the end of the experiment to avoid interference with
other compounds or omit them from the experiment altogether.
4.1.6 Evaluating results
At the simplest level, LMW screens are evaluated by comparing the binding
responses corrected for analyte molecular weight, and setting a cut-off level
above which binding is regarded as significant. The cut-off may be determined
by comparison with the response obtained from known non-binders (negative
controls) or set to some level chosen in relation to the distribution of response
values.
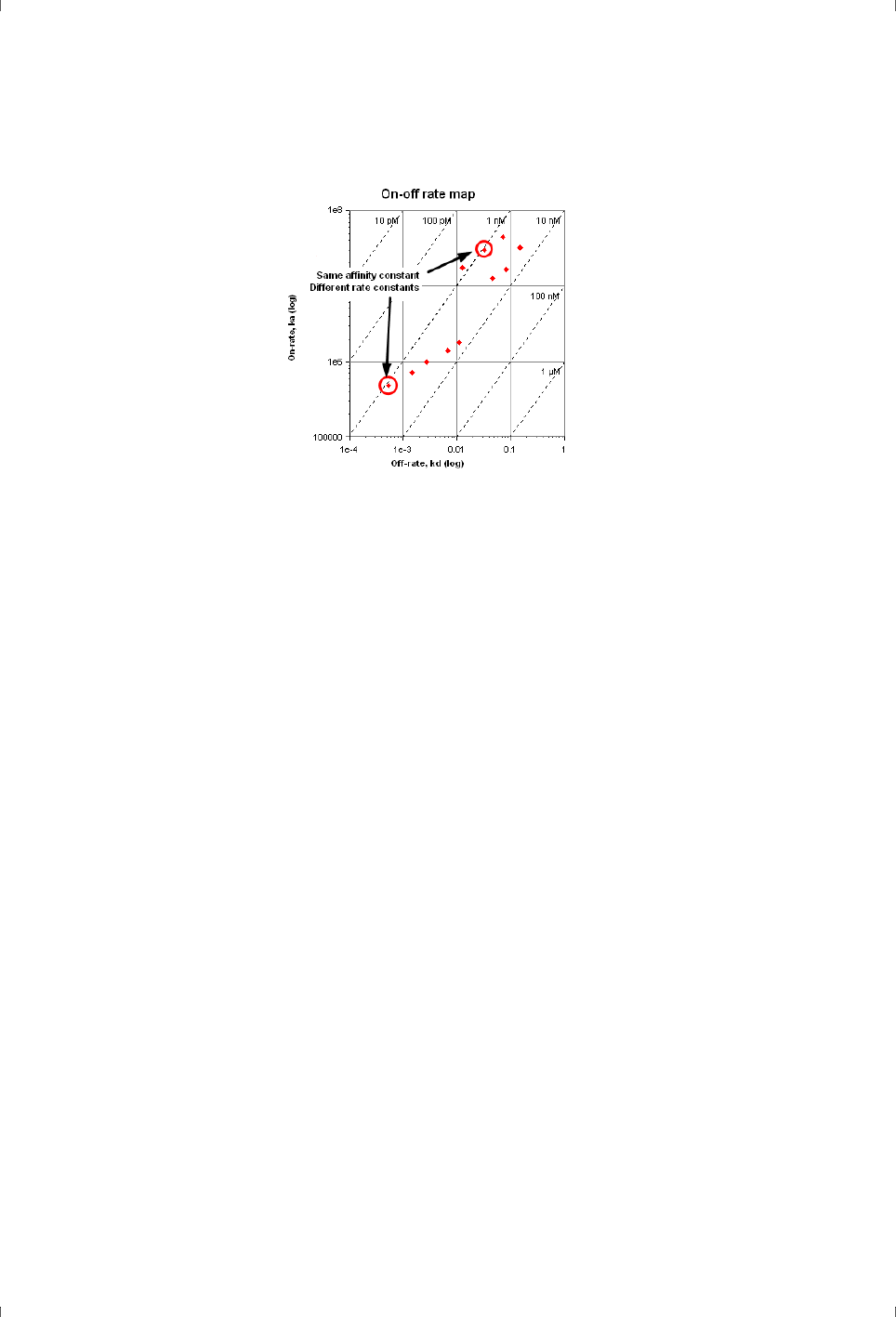
A valuable way to compare kinetic properties is to plot the association rate
constant against the dissociation rate constant on logarithmic axis. Diagonal
lines in this plot represent constant affinity, so that compounds that lie on the
same diagonal but are separated from each other have the same affinity but
different kinetics (Figure 4-2).
Biacore Assay Handbook 29-0194-00 Edition AA 35
Screening and detecting binding partners 4
Figure 4-2. Plotting association rate constant k
a
against dissociation rate constant k
d
(here on logarithmic scales) creates a plot where the affinity is represented by diagonal
lines. Compounds on the same diagonal have the same affinity but differ in kinetics.
Fragment screening benefits from additional steps in experimental design and
evaluation, to identify and characterize interesting fragments. In summary, the
recommended approaches are:
• Clean screen, aimed at rapidly identifying and eliminating undesirable
“sticky” compounds that show persistent binding to the surface and can
disturb subsequent analysis cycles.
• Binding level screen, aimed at providing a rapid overview of the compound
library, identifying compounds with binding levels above a defined cut-off.
• Affinity screen, aimed at verification of binding and affinity ranking of
fragments.
These approaches are described in more detail in the handbooks for Biacore
systems that support fragment screening.
4.2 Antibody screening
4.2.1 Goals
The goal of antibody screening is to identify cell clones that produce antibodies
appropriate for the purpose (usually biopharmaceuticals or biochemical tools).
Screening needs to be rapid so that clones can be selected before they grow to
unsuitable cell densities. Obtaining kinetic information early in the screening
process is often a significant advantage. Information that is generally sought
includes:

4 Screening and detecting binding partners
4.2 Antibody screening
36 Biacore Assay Handbook 29-0194-00 Edition AA
• Which clones produce antibodies with suitable specificity
• Which antibodies have kinetic and/or affinity properties suitable for the
purpose
• Which of these clones produce antibodies in sufficient amounts
4.2.2 Sensor surface preparation
Unlike LMW screening, expected response levels in antibody screening can be
quite high, so there is no need to use high levels of immobilized or captured
ligand. Typically, sensor surfaces are prepared with a generic anti-antibody
such as anti-IgG for which assay conditions (including regeneration) are known,
and binding characteristics are determined with an injection of antigen after
antibody capture. It is also possible to perform an antibody screen directly with
antigen attached to the surface as ligand, but this approach often involves
additional work to establish regeneration conditions.
4.2.3 Sample preparation
Samples for antibody screening are typically taken from cell culture supernatant
and analyzed directly without purification. Preliminary experiments with non-
producing clones to establish that there is no significant binding of non-
antibody components can simplify interpretation of the results.
4.2.4 Buffers
HBS-EP+ (Hepes-buffered saline with 0.3 mM EDTA and 0.05% Surfactant P20,
available from GE Healthcare) or similar is recommended as running buffer.
Samples may be diluted if required in the same buffer. Precise matching of
sample and running buffer is not practicable for screening work.
4.2.5 Analysis conditions
Flow rate
Antibody screening by capture on a generic capturing molecule can be run at
low flow rates (typically 10 µl/min) if sample consumption is an issue.
Characterization of the antibody-antigen interaction, whether performed
directly on immobilized antigen or by a secondary injection over captured
antibody, should be run at moderate flow rates (typically 30 µl/min) to reduce
mass transfer limitations (see Section A.1).
Contact time
Contact times should be sufficient to give confidently measurable response
levels without compromising screening throughput. Contact times of 1 to 2
minutes are usually sufficient.
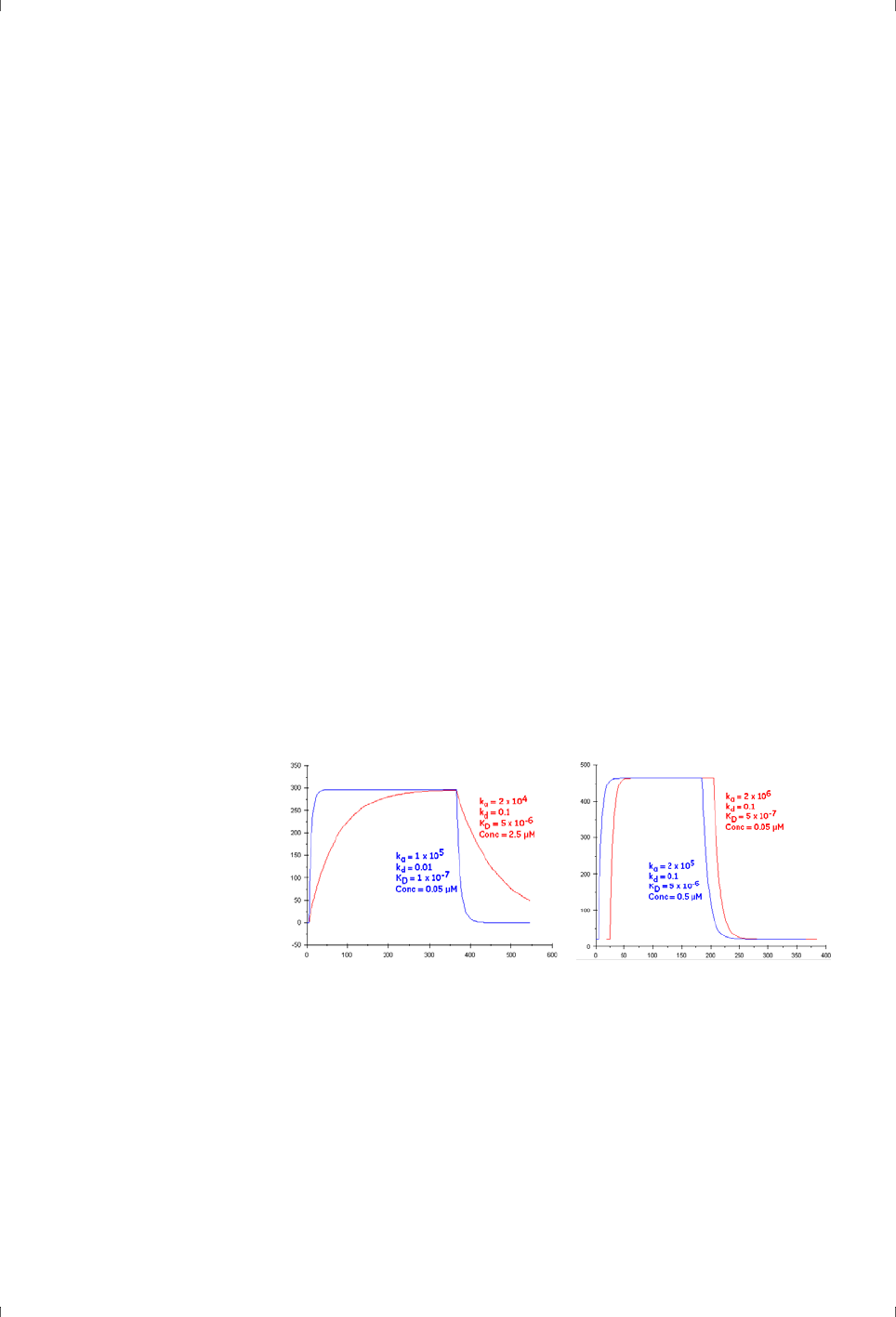
Biacore Assay Handbook 29-0194-00 Edition AA 37
Screening and detecting binding partners 4
Regeneration
Regeneration conditions for antibody screening kits from GE Healthcare are
specified in the respective Instructions for Use. For assay development using
custom antibodies, regeneration at low pH (glycine-HCl, pH 1.5 to 3) is usually
effective.
Report points
For screening, place a report point shortly after the end of the sample injection.
This will avoid any bulk refractive index differences between sample and
running buffer.
Antibody characterization based on complex stability uses report points placed
early and late in the dissociation phase of the antibody-antigen interaction.
4.2.6 Evaluating results
The response reached in the first step of the screening gives an indication of the
relative concentration of antibodies in the sample (according to the specificity
of the capturing molecule). Responses achieved from injection of antigen reflect
both the antigenic specificity and the binding characteristics of the captured
antibody. Note however that evaluation based on report points alone does not
give unambiguous information: a given response may be obtained from a
combination of high concentration and slow binding or low concentration and
fast binding. It may be possible to resolve these alternatives by visual inspection
of the sensorgrams, but if the binding and dissociation events are rapid both
situations may give closely similar sensorgrams (see Figure 4-3).
Figure 4-3. Sensorgram shape can in some cases distinguish weak and strong binders
that reach the same response level as a result of different concentrations (left). In other
cases, depending on the relative values of the rate constants and concentrations, the
sensorgrams may be identical. These sensorgrams are simulated: those in the right-hand
panel are displaced slightly from each other on the time axis.
Frequently, the parameter of most interest in choosing between antibody clones
is the stability of the antibody-antigen complex, which is largely governed by the
dissociation rate. Comparison of the response early and late in the dissociation
phase can give a rapid indication of the relative dissociation rates.

4 Screening and detecting binding partners
4.3 Immunogenicity testing
38 Biacore Assay Handbook 29-0194-00 Edition AA
4.3 Immunogenicity testing
4.3.1 Goals and challenges
Immunogenicity testing answers the question of whether administered drugs
provoke an immune reaction in the recipient that can compromise the efficacy
of the drug. At a simplistic level, this application is analogous to screening for
antibodies in serum, and recommendations for screening apply also to
immunogenicity testing, using immobilized drug as ligand. However, the
simultaneous presence of the drug and circulating anti-drug antibodies (ADAs)
in serum presents a special challenge: ADAs in complex with drug will not be
detected in an assay based on binding to the drug.
ADAs are released from drug complexes under acidic conditions (pH 1 to 2), and
can be detected if the sample is neutralized and the assay is performed
immediately, before the complexes have had time to re-form fully. This
approach is implemented in some Biacore systems, using a specially designed
IFC that mixes acidified samples with neutralizing solution immediately before
injection over the sensor surface. Details of the approach are described in the
manuals for systems that support immunogenicity testing (e.g. Biacore T200
Immunogenicity Handbook).

Biacore Assay Handbook 29-0194-00 Edition AA 39
Kinetics and affinity measurements 5
5 Kinetics and affinity measurements
5.1 Approaches
Determination of both kinetics and affinity rely on measurements over a range
of analyte concentrations. Kinetic (rate) constants are obtained by analyzing the
sensorgram shape in relation to a mathematical model of the interaction
mechanism: affinity constants are obtained from the steady state binding
levels. Affinity constants can also be calculated from the ratio of the rate
constants. Details of the data analysis principles are given in Appendix A.
5.1.1 Single- and multi-cycle kinetics
There are two approaches to determination of kinetics and affinity, both based
on determination of binding data at several (typically 5) analyte concentrations
(see Figure 2-2):
• multi-cycle kinetics, where each analyte concentration is injected in a
separate analysis cycle, and the sensor surface is regenerated between
cycles. Since data from the different concentrations is evaluated together,
it is important that regeneration is carefully optimized so that the surface
properties are constant between cycles.
• single-cycle kinetics, where analyte concentrations are injected
sequentially in a single cycle, with no regeneration between injections.
Both approaches have been shown to give equivalent results in test systems,
and both can be used for steady-state affinity measurements if the sample
injection time is long enough for steady state to be reached.
The single-cycle approach requires less time for a complete analysis, and also
benefits from the lack of requirement for optimized regeneration. This can
significantly reduce the demands on assay development, and permits kinetic
determinations for interactions where regeneration conditions are not
available. However, the cycle time is longer in single-cycle kinetics, so the
approach is more sensitive to drift such as dissociation of ligand in a capture
assay.
5.1.2 2-over-2 kinetics
Normally, all samples are analyzed over a single sensor surface, providing
identical ligand conditions for each analyte concentration. In Biacore A100 and
Biacore 4000 systems, however, parallel analysis over multiple ligand spots in
each flow cell permits a technique called 2-over-2 kinetics. The name refers to
analysis of two analyte concentrations over two ligand densities, and can be
performed in a single cycle in Biacore systems which can support multiple

5 Kinetics and affinity measurements
5.2 Sensor surface preparation
40 Biacore Assay Handbook 29-0194-00 Edition AA
ligand densities in each flow cell. Combining the data from two ligand densities
compensates for the lack of analyte concentrations, and the approach can
provide reliable kinetic data from experiments that maintain a moderately high
sample throughput.
5.1.3 Comparative estimates of kinetics and affinity
Comparison of relative interaction kinetics can be sufficient for some
applications. At the simplest level, this can be achieved by plotting the ratio of
response values early and late in the sample injection (for association rates) and
in the dissociation phase (for dissociation rates), using a single analyte
concentration. This simplified approach is sometimes used in information-rich
screening work, where full determination of kinetics and/or affinity is not
practicable for throughput considerations.
5.2 Sensor surface preparation
Amount of immobilized ligand
In evaluation of kinetic and affinity constants, the concentration of ligand is
expressed in resonance units (RU) so the amount of immobilized ligand does not
have to be known exactly (see Appendix A). The immobilization level is however
important for the quality of the results.
For kinetic analysis, the amount of immobilized ligand should be kept low, so
that the maximum response from analyte binding is typically in the region of 30
to 50 RU or lower. There are two main reasons for this:
• Low ligand levels help to reduce limiting effects of mass transfer. Mass
transfer refers to the diffusion-controlled supply of analyte molecules to
the surface from bulk solution: if transfer is slow in relation to the
association rate, the observed binding will be a measure of the diffusion
process and not of the interaction rates. Mass transfer is discussed in
more detail in Appendix A.
• Low ligand levels also help to minimize artifacts that can arise from
crowding of molecules on the surface. Crowding effects are difficult to
quantitate but can give rise to unnecessarily complex binding behavior.
For measurement of steady-state affinity, mass transfer considerations are not
relevant and the amount of immobilized ligand can be higher than for kinetic
analysis.
Reference surface
Kinetic and affinity measurements should always use reference-subtracted
data. The reference surface may be untreated, activated and deactivated, or
prepared with an inactive protein to mimic the physical properties of the active
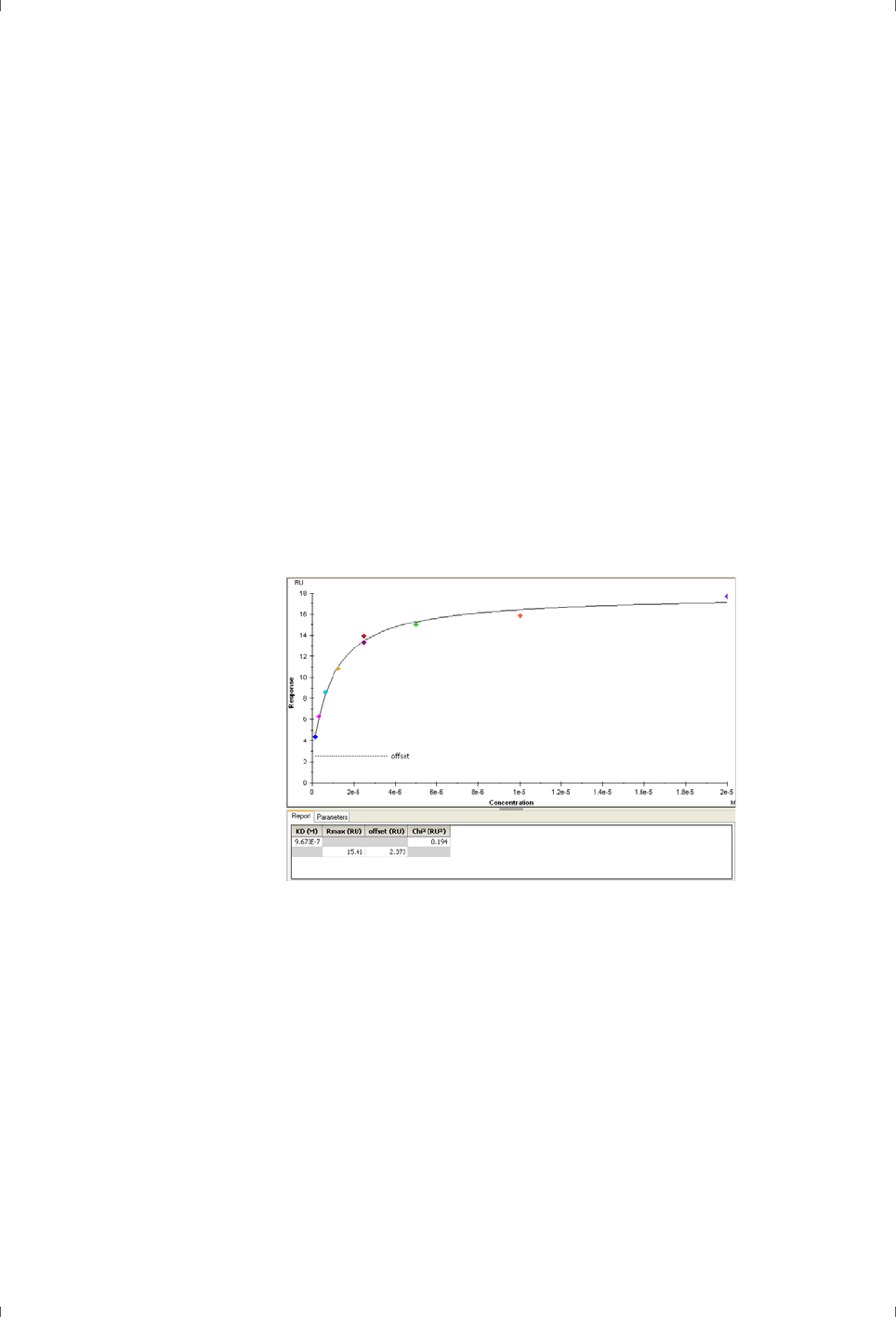
Biacore Assay Handbook 29-0194-00 Edition AA 41
Kinetics and affinity measurements 5
surface. These alternatives are discussed in more detail in the Biacore Sensor
Surface Handbook.
5.3 Buffers
The same buffer must be used for sample and running buffer, so that
association (during sample injection) and dissociation (after the end of the
injection) occur in the same environment. Moreover, evaluation of kinetics is
more robust if the sample and running buffer are matched in refractive index, so
that bulk shifts at the beginning and end of the sample injection are kept as
small as possible.
Buffer matching is less important for affinity measurements since evaluation is
based only on response values measured during the sample injection. Bulk
response contributions will however introduce an offset from the x-axis in the
plot of response against analyte concentration (Figure 5-1). It is important that
bulk effects do not vary between samples so that the offset is constant.
Figure 5-1. Plot of response against analyte concentration evaluated with offset to allow
for bulk effects.
5.4 Sample preparation
Preparation of samples can be a critical factor in the quality of kinetic
measurements. Ideally, both ligand and analyte should be pure and
homogeneous, to simplify the interpretation of the results as far as possible.
Using a capturing approach for attaching ligand provides on-chip purification:
however, it is essential for careful work that the capturing interaction is
sufficiently stable so that ligand does not dissociate during the sample analysis.
It is important that non-specific binding of sample components to the sensor
surface is kept to a minimum. It is also important that the analyte is

5 Kinetics and affinity measurements
5.5 Sample concentrations
42 Biacore Assay Handbook 29-0194-00 Edition AA
homogeneous and non-aggregated since differently sized binding species will
give different responses. Use non-denaturing analysis methods to check
analyte homogeneity.
Samples may be diluted into running buffer from stock solution if the dilution
factor is sufficiently high so that residual stock solution components can be
ignored. Alternatively, samples may be prepared using buffer exchange
techniques.
5.5 Sample concentrations
Evaluation of association rate constants and affinity constants from the
experimental data is directly dependent on the values provided for analyte
concentrations (see below for details). For this reason, it is important for
obtaining correct results that the analyte concentration is determined as
accurately as possible. Ideally, this should be the concentration of analyte that
is capable of interacting with ligand, which may not be the same as the total
concentration (if for example the analyte is only partially active). Calibration-free
concentration analysis (see Section 2.3.2) provides an approach to
concentration measurement that is directly related to the amount of analyte
that can bind to the ligand, and is recommended in critical measurements. If
there are doubts about the validity of the analyte concentration values, the
reported values for the association rate constant and affinity constant should
be treated with due caution. The dissociation rate constant is not dependent on
the analyte concentration.
Analyte concentration range
In principle, kinetic constants can be obtained from analysis at a single analyte
concentration, but analysis over several concentrations (5 to 8 for multi-cycle
kinetics, up to 5 for single-cycle kinetics) gives a better foundation for
discovering deviations from the simplest interaction models and for revealing
concentration-dependent artefacts (for example analyte aggregation at
increasing concentrations). Ideally, concentrations should cover a wide range
centered on the value for the dissociation equilibrium constant K
D
(for example,
for an interaction with K
D
= 10 nM, concentrations from 1 to 100 nM are
appropriate). In practice, the K
D
value is often not known, and the range of
concentrations that can be used may be limited by availability or solubility of the
analyte. A concentration range that gives a good spread of sensorgrams is
usually adequate (Figure 5-2).
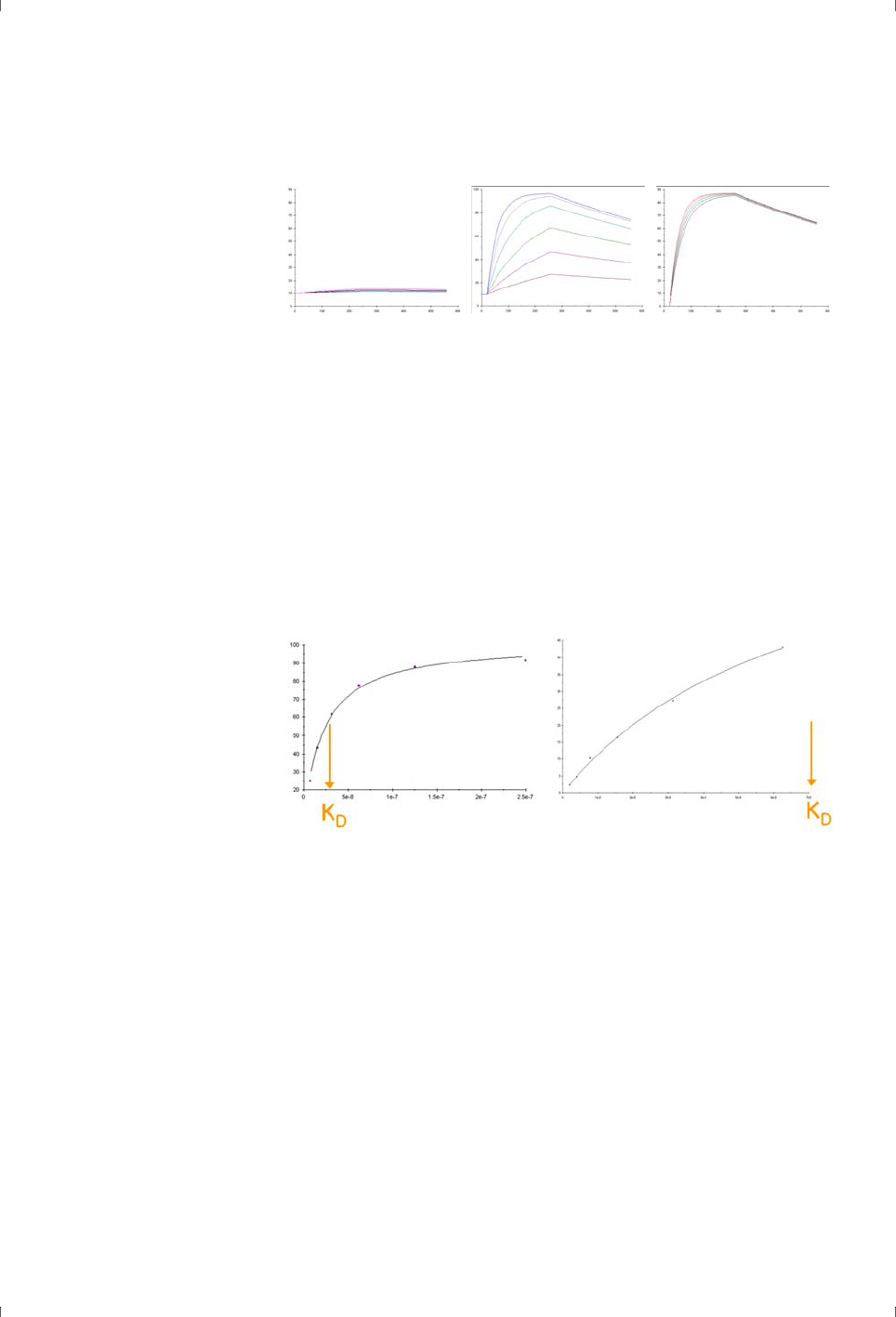
Biacore Assay Handbook 29-0194-00 Edition AA 43
Kinetics and affinity measurements 5
Figure 5-2. Analyte concentrations for kinetic analysis should cover a wide range (center
panel). If the concentrations are too low (left panel) or too high (right panel) the
sensorgrams may be tightly grouped and evaluation will be less robust. The vertical scale
for these simulated sensorgrams is the same in all three panels.
For affinity determinations, each analyte concentration gives rise to a single
point on the experimental data plot, in contrast to kinetic analysis where each
analyte concentration provides a full time-course of the interaction. It is
therefore important to use a sufficient number of analyte concentrations in the
assay (the recommended minimum is 5). In addition, the concentration range
should extend to more than twice the affinity constant (K
D
) value. If these
conditions are not fulfilled, there will be insufficient data in the plot of R
eq
against
C for confident analysis. Conversely, a reported K
D
value higher than half the
highest analyte concentration should be treated with care (Figure 5-3).
Figure 5-3. The analyte concentration range for steady state affinity determination
should cover more than twice the K
D
value (left). Do not trust reported affinity constants
that are higher than half the highest analyte concentration (right).
Analyte concentration values
The association rate constant for a simple binary interaction has units of M
-1
s
-1
(see Appendix A), and molar analyte concentrations must be supplied to the
software for kinetic evaluation. Similarly, evaluation of steady state affinity
requires analyte concentrations as input. It is important that analyte
concentrations are correct if correct values for rate constants are to be
obtained. Incorrect values for analyte concentration will lead directly to
corresponding errors in the association rate constant k
a
.
Kinetic measurements rely on analysis over a range of analyte concentrations,
usually prepared by serial dilution from a stock solution. Properly calibrated
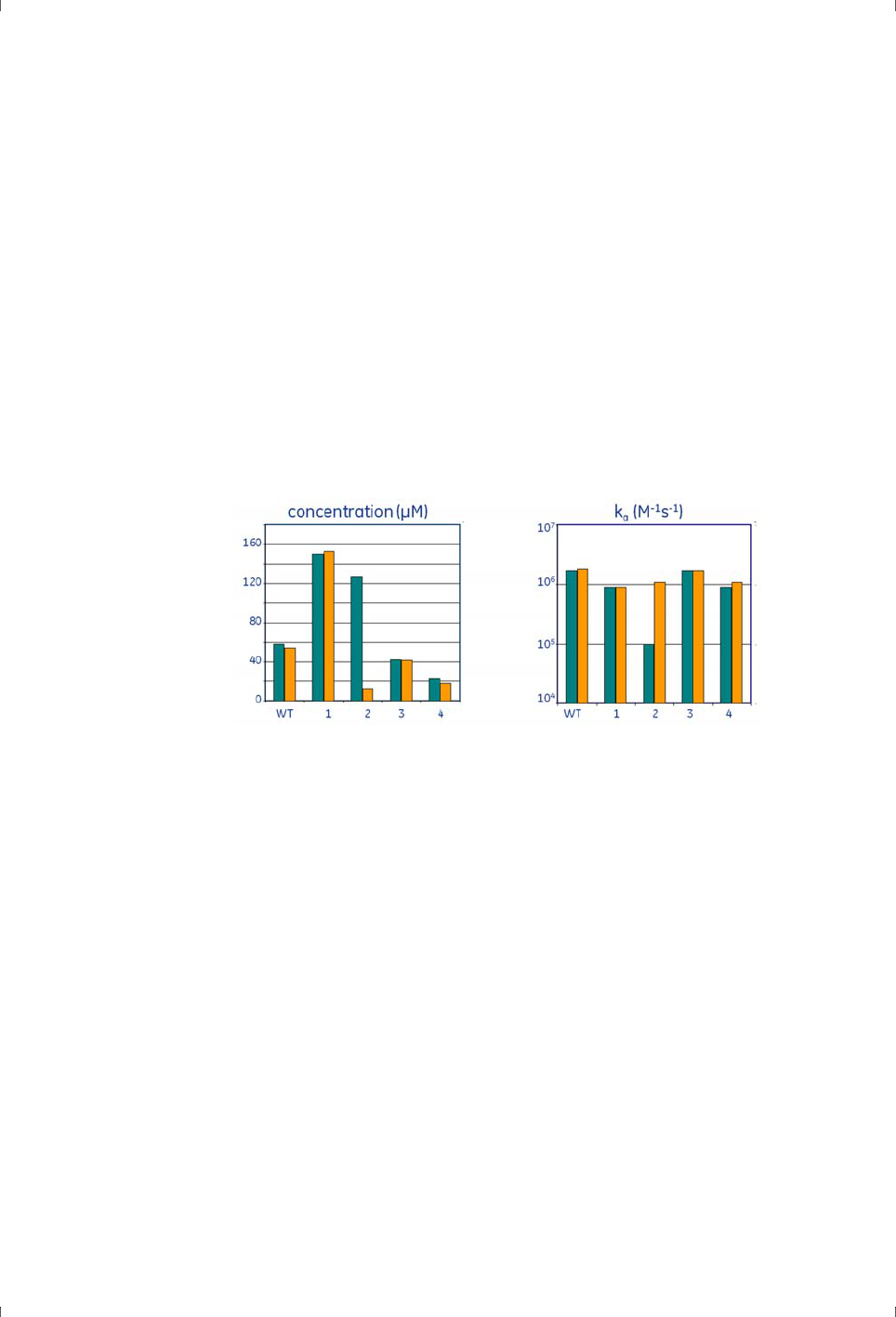
5 Kinetics and affinity measurements
5.5 Sample concentrations
44 Biacore Assay Handbook 29-0194-00 Edition AA
equipment and well-designed sample preparation protocols are essential if the
concentrations are to be correct (see Section 3.5.
In addition to accurate sample preparation, correct values for the stock solution
concentration are important. The ideal value is the concentration of analyte
that is capable of binding to the surface-attached ligand: this may not
necessarily be the same as the total analyte concentration. If half of the analyte
molecules are in a conformation that cannot bind, the total concentration will
be twice the value that should be used for evaluation of binding kinetics. If the
sample is amenable to concentration determination by CFCA, this is the value
that should ideally be used: it represents the concentration capable of
interaction and is independent of calibration considerations. Figure 5-4
illustrates an example of the importance of using the correct analyte
concentration.
Figure 5-4. Importance of using the correct analyte concentrations. The experiment
compared the kinetics of wild type and four mutants of a papain inhibitor binding to
papain. Using concentrations obtained from A
280
measurements (green bars), mutant
number 2 had an association rate constant about one order of magnitude lower than the
wild type and the other mutants. However, using concentrations obtained by CFCA
(orange bars), association rate constants obtained were similar for all variants of the
inhibitor.

Biacore Assay Handbook 29-0194-00 Edition AA 45
Epitope mapping 6
6 Epitope mapping
Epitope mapping refers to determination of the relative topography of binding
sites (usually for antibodies) on a target molecule or antigen. Antibodies that
bind to epitopes that are distinct from each other can bind simultaneously to the
antigen, while antibodies directed against the same or interfering epitopes
show mutually exclusive binding.
The commonest approach to epitope mapping is to test simultaneous binding
ability in a pair-wise matrix. In an alternative approach, peptides representing
structural epitopes on the antigen are tested for inhibition of antibody binding.
Both methods are amenable to use with Biacore systems.
6.1 Pair-wise binding
6.1.1 Principle
The basic principle of pair-wise mapping studies in Biacore systems is
straightforward: one antibody is attached to the surface, antigen is bound to the
antibody, and a second antibody is tested for simultaneous binding ability. In
practice, the assay set-up involves additional steps (see Figure 6-1):
• The first antibody to the sensor surface is conveniently attached to the
sensor surface by capturing on a generic antibody such as rabbit anti-
mouse immunoglobulins for mouse antibodies. This allows multiple
antibody pairs to be tested using the same sensor surface. Permanent
attachment is less suitable since the first antibody cannot then be
changed without preparing a new sensor surface.
• It is important to ensure that no antibody-capturing sites on the surface
are available to bind the second antibody when it is injected. Remaining
capture sites are therefore blocked with an injection of blocking antibody
(an antibody that does not bind to the antigen being tested).
• Antigen is then injected and binds to the first antibody.
• The antigen is followed by an injection of second antibody to test for
simultaneous binding of the two antibodies.
• The surface is regenerated to remove all components except the
permanently attached capturing antibody, in preparation for testing the
next antibody pair.
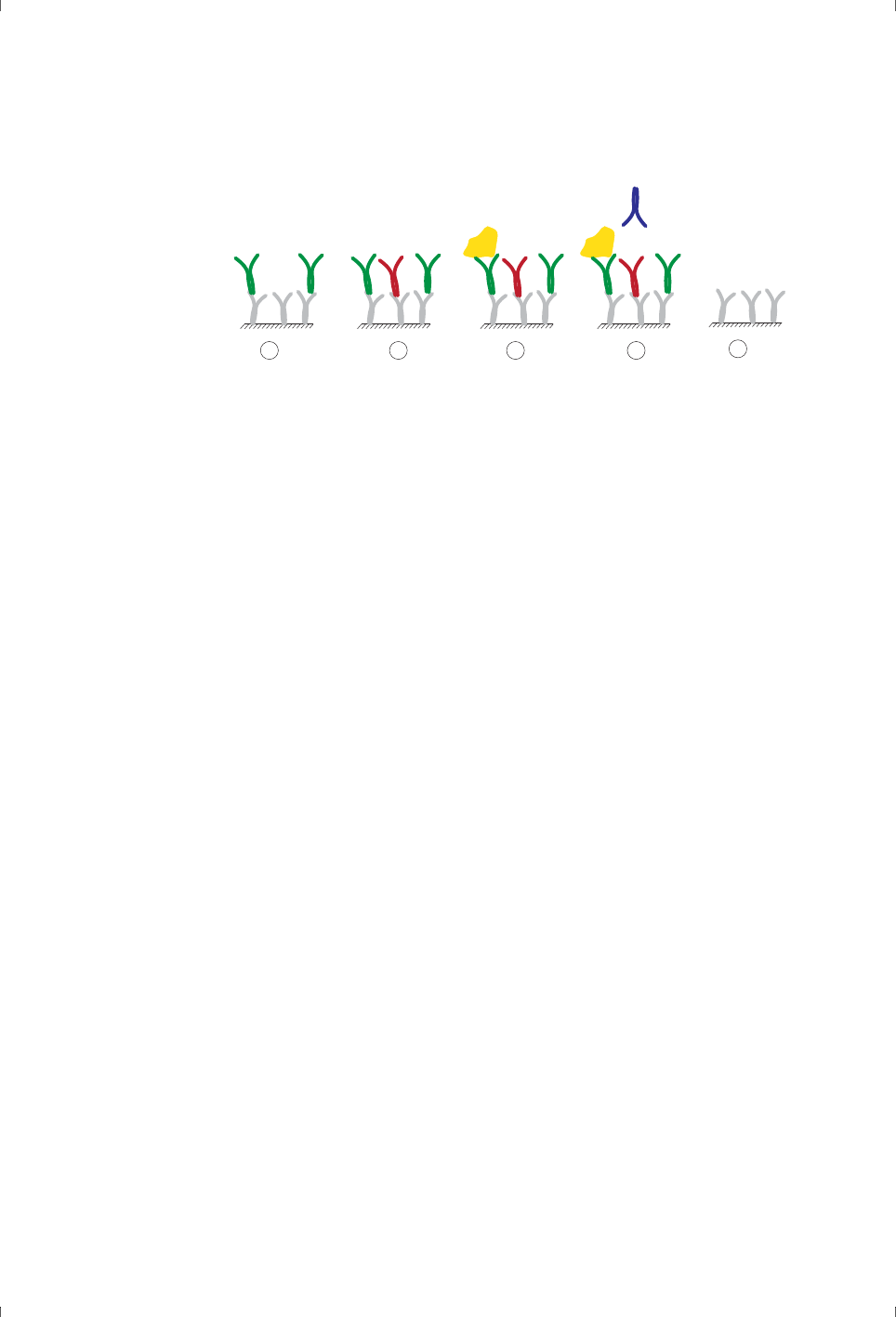
6 Epitope mapping
6.1 Pair-wise binding
46 Biacore Assay Handbook 29-0194-00 Edition AA
Figure 6-1. Main steps in a pair-wise epitope mapping experiment.
1. Capture first antibody.
2. Block remaining capture sites.
3. Inject antigen.
4. Challenge with second antibody.
5. Regenerate in preparation for the next antibody pair.
A panel of n antibodies contains n(n-1)/2 unique pairs of non-identical
antibodies (for example 5 antibodies give 10 pairs). It is recommended to test all
combinations in the pair-wise matrix:
• Pairs where the first and second antibody are identical provide a check on
the assay performance. If simultaneous binding is observed this may
indicate that the antigen contains multiple copies of the epitope, which
may invalidate the mapping with respect to that epitope.
• Pairs where the roles of first and second antibody are reversed provide a
control for unexpected effects such as conformational changes induced
by antibody binding. Ideally, the results should be independent of the order
in which antibodies are tested, but this is not always the case in practice.
6.1.2 Evaluation
The label-free detection in Biacore systems offers advantages over label-based
methods for epitope mapping, in that all steps in the binding sequence are
monitored with the same technique. It is easy to relate the observed level of
antigen binding to the level of first antibody, and the level of second antibody to
that of antigen. This addresses a potential issue in interpreting the results, in
that the absence of second antibody binding may result either from interference
between the epitopes or from poor binding of the antigen to the first antibody.
This cannot be resolved in methods where antigen binding is not monitored
directly.
Epitope mapping applications are explicitly supported in Biacore 4000, with
methods for both set-up and evaluation of pair-wise mapping experiments
(described in detail in the Biacore 4000 Software Handbook). Evaluation is
divided into five distinct steps, summarized below. Similar principles can be
applied in evaluating pair-wise mapping results from other Biacore systems.
?
1
2 3 4
5

Biacore Assay Handbook 29-0194-00 Edition AA 47
Epitope mapping 6
Report points for each evaluation step are set just after the end of the respective
injections.
First antibody ranking
This step presents captured levels of the first antibody. The amount of first
antibody is fundamental in determining the possible response levels in
subsequent steps, and may vary between antibodies. A cut-off boundary can be
set to represent the minimum acceptable level: below this level, the amount of
captured first antibody is judged to be too low for reliable measurement in
subsequent steps, and the antibody pairs represented by these points are
excluded from the evaluation.
Antigen ranking
Even with sufficient first antibody captured, binding of antigen may vary, and
results for binding of the second antibody will be inconclusive if the level of
antigen bound is too low. As for first antibody binding, a cut-off boundary can
be set to exclude points with response values below an acceptable limit. The
binding data may be shown as relative response values or as percent of
expected binding (based on the amount of first antibody captured and the
relative sizes of antibody and antigen).
Second antibody ranking
The second antibody ranking displays the binding levels of second antibody,
either as relative response or as percent of expected binding (based on the
amount of first antibody captured and the relative sizes of antibody and
antigen). Here again a cut-off boundary can be set to differentiate between
positive and negative binding.
Dissociation fit
An additional component in the evaluation is estimation of the dissociation rate
of antibodies classed as positive binders in the second antibody ranking step.
This information is not essential for pair-wise mapping itself, although it can be
useful in judging the status of borderline results. However, it is often useful in the
context of antibody development for immunoassay reagents, and is obtained
without additional experimental design or set-up from determinations with
Biacore systems.
Result matrix
The results of pair-wise epitope mapping are collected in a matrix of binding
values, with first and second antibodies in columns and rows respectively. Cells
in the matrix are classed as positive (meaning independent epitopes) or
negative (interfering epitopes) on the basis of the cut-off boundary set for the
second antibody ranking step. If the cut-off boundaries have been judiciously
set in the first antibody and antigen ranking steps, the classification in the
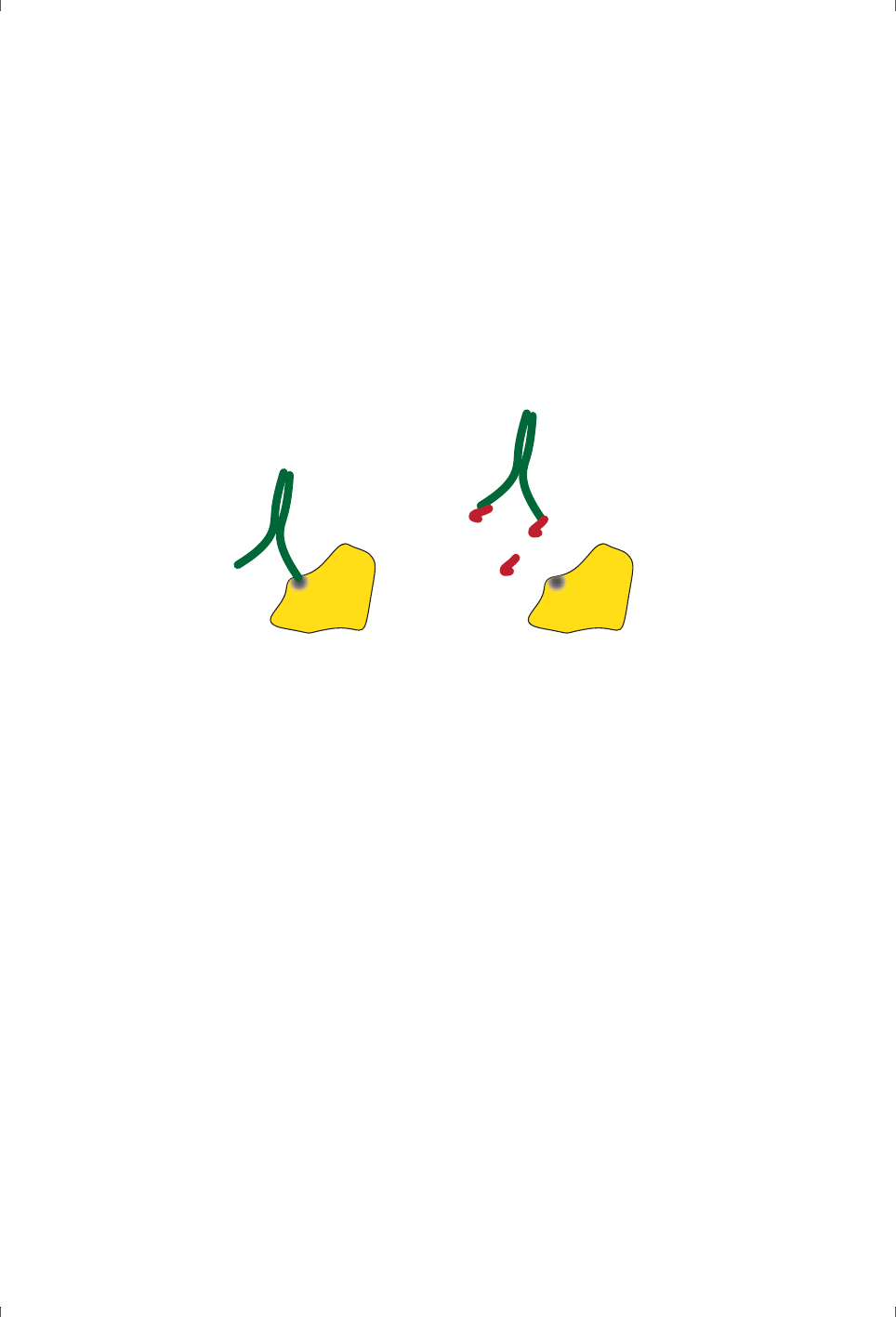
6 Epitope mapping
6.2 Peptide inhibition
48 Biacore Assay Handbook 29-0194-00 Edition AA
matrix reflects only those pairs where second antibody binding is potentially
reliable.
6.2 Peptide inhibition
Epitope mapping by peptide inhibition is based on the principle that a peptide
representing a structural epitope will inhibit binding to antigen of an antibody
directed against that epitope (Figure 6-2). Inhibition may be partial or complete
depending on how well the peptide is structurally congruous with the epitope.
Figure 6-2. Principle of epitope mapping by peptide inhibition principle. In the absence of
peptide (left), antibody can bind to the epitope. Added peptide that mimics the epitope
binds to the antibody (right) and blocks binding to the antigen.
An assay of this kind can be set up with either the antigen or antibody attached
to the surface. Antigen may be attached permanently and regenerated:
antibodies are captured to allow investigation of multiple antibodies using the
same sensor surface.
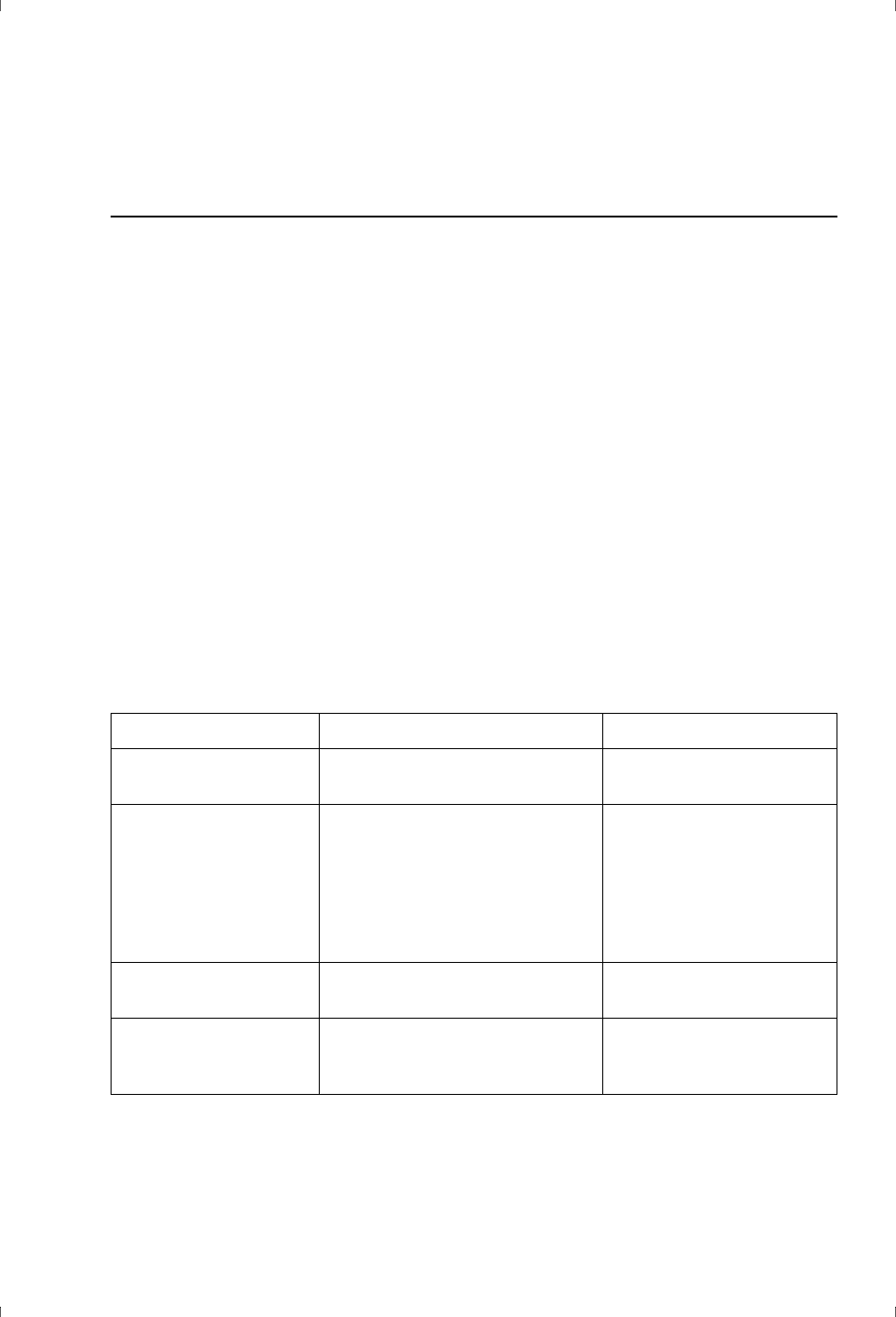
Biacore Assay Handbook 29-0194-00 Edition AA 49
Troubleshooting assays 7
7 Troubleshooting assays
This chapter considers the causes and suggested solutions to common
problems with Biacore-based assays. The material is divided into 4 sections:
• Ligand attachment
•Analyte binding
• Dealing with non-specific binding
• Unexpected sensorgram shape
7.1 Troubleshooting ligand attachment
This section deals with the problem that the amount of ligand attached to the
surface is too low for the experimental purpose.
7.1.1 Covalently attached ligand
A schematic sensorgram for immobilization of a macromolecular ligand on
Sensor Chip CM5 is illustrated in Figure 3-3. While details differ according to the
chemistry and the specific molecules involved, there are some common
features that can help in diagnosing problems with ligand immobilization.
Symptom Cause Remedy
Poor response increase
during ligand injection
Immobilization pH unsuitable Optimize immobilization pH
Poor response increase
during ligand injection
Too high ionic strength in
immobilization buffer
Use low ionic strength (10 mM
monovalent cations).
Ligands kept in stock solution
with physiological ionic
strength should be diluted at
least 10-fold or otherwise
desalted.
Poor response increase
during ligand injection
Ligand concentration too low Test higher concentrations
Ligand binds during
injection but is not
retained
Ligand lacks accessible target
groups for immobilization chemistry
Try a different chemistry or
use capturing
7 Troubleshooting assays
7.1 Troubleshooting ligand attachment
50 Biacore Assay Handbook 29-0194-00 Edition AA
7.1.2 Captured ligand
Ligand capture by high affinity binding to an immobilized protein involves
attachment of the capturing molecule to the sensor surface followed by binding
of the ligand to the capturing molecule. From the viewpoint of troubleshooting,
the first step is equivalent to immobilizing a ligand, while the second is
equivalent to binding analyte.
There are however some specific capture situations supported by ready-to-use
sensor chips that can present individual problems in ligand attachment.
Laboratory protocols with recommended conditions are available from GE
Healthcare.
• Capture of biotinylated ligands on Sensor Chip SA or with the Biotin
Capture Kit requires that excess biotinylation reagents are rigorously
removed from the ligand solution (suitably be gel filtration on microspin
columns). Any residual biotinylation reagents in the solution will compete
with ligand for attachment to the surface and reduce capture efficiency.
Removal by two cycles of desalting is recommended.
• Capture of histidine-tagged ligands by metal chelation on Sensor Chip NTA
requires that buffers are free from chelating agents (EDTA is recommended
for ligand removal). The efficiency and stability of capture varies
considerably with the number, length and location of the polyhistidine tags
on the ligand molecule, but generally improves with reduced ligand levels.
If capture on Sensor Chip NTA is unsatisfactory, you may want to try using
longer or multiple polyhistidine tags, or capture the ligand on anti-histidine
antibodies instead.
Ligand binds during
injection but is not
retained
Buffer components such as Tris or
additives such as sodium azide
compete with the ligand for
immobilization
Make sure the buffer is
appropriate for the
immobilization chemistry.
Immobilization level low Old EDC/NHS solutions Use fresh EDC and NHS
solutions.
One or more injections do
not show the
characteristic response
Solutions wrongly placed in
autosampler
Check the Rack Positioning
information in the software
for the immobilization
procedure.
Symptom Cause Remedy

Biacore Assay Handbook 29-0194-00 Edition AA 51
Troubleshooting assays 7
7.2 Troubleshooting analyte binding
7.2.1 Analyte capacity too low
Estimate the activity of the attached ligand in terms of the measured analyte
binding capacity in relation to the theoretical value (Section 3.2.3). If analyte
binding is unacceptably low, the ligand may have lost activity in the low ionic
strength, low pH immobilization buffer, been inactivated by the covalent
attachment chemistry, or had low activity in the starting material. Try using a
different chemistry or switch to a capturing approach.
Occasionally, the attachment chemistry may inactivate the ligand by direct
modification of the binding site. In such cases, attaching the ligand in the
presence of a reversible binder that blocks the active site can help to preserve
activity. It is of course essential that the binder itself is not susceptible to the
attachment chemistry. This approach has proved successful in work with some
target molecules for low molecular weight analytes.
7.2.2 Analyte binding too high
Analyte binding capacity that exceeds the theoretical maximum value usually
indicates either that the binding stoichiometry is more than expected (either
because there are multiple binding sites on the ligand or because the analyte is
aggregated), or that the analyte is binding indiscriminately to the ligand or the
surface matrix. In the former case, it is in principle possible to saturate the
surface with sufficiently high analyte concentrations, giving an experimental
value for the observed stoichiometry. In the case of indiscriminate binding, the
surface can often not be saturated and the binding level continues to increase
with analyte concentration even at very high concentrations.
7.3 Dealing with non-specific and unwanted binding
7.3.1 Non-specific binding
Non-specific binding refers to indiscriminate binding of the analyte or other
component in the sample to either the ligand or the sensor surface matrix. Non-
specific binding can give false positive results in screening assays and incorrect
results in concentration assays. In determination of kinetics and affinity, non-
specific binding generally gives results that do not fit to an interaction model.
Typically, non-specific binding of analyte gives response levels that continue to
increase with increasing sample concentration, instead of reaching a saturation
plateau.
Binding to the sensor surface matrix is usually revealed by a binding response
on the reference surface. Subtraction of the reference response is not sufficient
to correct for non-specific binding since it is difficult to establish that binding to

7 Troubleshooting assays
7.4 Unexpected sensorgram shapes
52 Biacore Assay Handbook 29-0194-00 Edition AA
the reference surface and the non-specific component of binding to the active
surface are equivalent (high ligand levels can mask dextran binding sites and
reduce non-specific binding to the surface matrix). Non-specific binding of this
kind can often be reduced by addition of soluble dextran to the samples, to
compete for binding to the dextran on the sensor surface. Soluble dextran is
available for this purpose from GE Healthcare as NSB Reducer.
Non-specific binding to the ligand does not give a response on a reference
surface that lacks ligand but usually shows as non-saturatable binding to the
active surface. Binding of non-analyte components may be an issue with
complex samples such as cell extracts or serum. General principles for
minimizing this kind of binding are to use physiological or higher ionic strength
in the sample and running buffer and to include detergent if this does not
interfere with the assay. If these measures are not sufficient, choose a different
ligand (for example a different clone for monoclonal antibody ligands).
Sometimes the analyte binds promiscuously to the ligand (for example, many
low molecular weight compounds bind with low affinity to multiple sites on
serum albumin, giving behavior that resembles non-specific binding). In such
cases, it may be possible to obtain useful information about high-affinity
binding sites from sensorgrams obtained at relatively low sample
concentrations, where low affinity binding does not dominate the observed
response. Adjustment of buffer conditions for the assay can also help to reduce
the effect of promiscuous binding.
7.3.2 Unwanted binding
Unwanted binding refers to the specific binding of non-analyte components to
the ligand, or binding of the analyte to non-ligand components such as a
capturing molecule on the surface. This kind of interference is best dealt with by
careful assay design. For example, avoid antibody subclasses that bind
complement factors for measurements in serum, or use Fab fragments that lack
the complement-binding sites.
7.4 Unexpected sensorgram shapes
This section considers interpretation and correction of unexpected sensorgram
behavior. There will always be individual differences in sensorgram shape
according to interaction characteristics, but the following general
characteristics are “expected” with due allowance for bulk refractive index
effects:
• The baseline should be stable within the analysis cycle. Minor baseline
shifts between cycles may be acceptable.
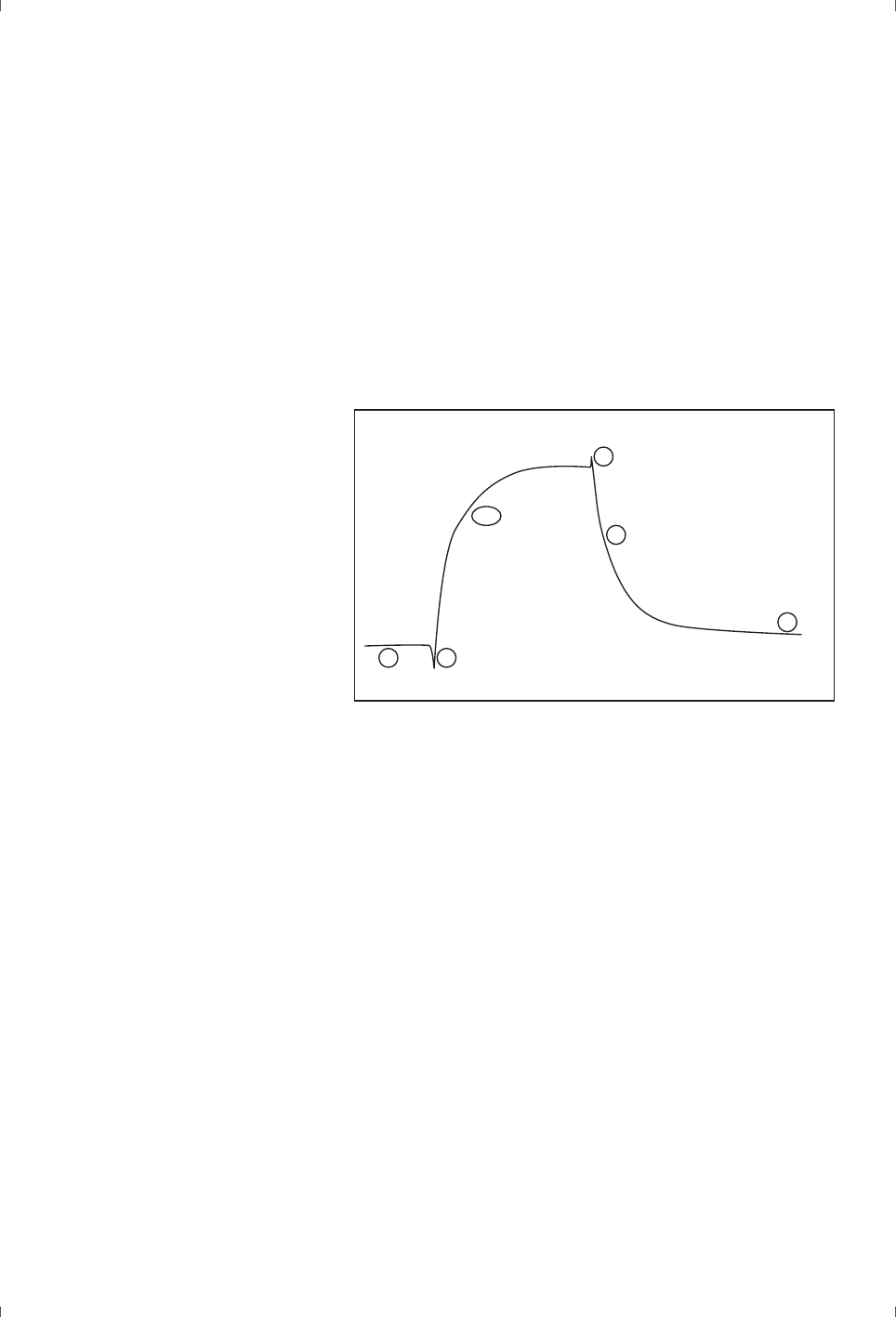
Biacore Assay Handbook 29-0194-00 Edition AA 53
Troubleshooting assays 7
• The response should not decrease progressively during sample injection
(with the exception of bulk shifts resulting from refractive index
differences).
• The response should not increase progressively during dissociation (with
the exception of bulk shifts resulting from refractive index differences).
• Reference-subtracted response overshoot at the start and end of sample
injection should be small.
• The response should not go below the baseline level during dissociation.
Figure 7-1. Generalized features of the “expected” shape of reference-subtracted
sensorgrams.
1: stable baseline.
2: positive response during sample injection.
3: increasing or stable response during injection.
4: decreasing response during dissociation
5: small reference subtraction spikes at the start and end of injection
6: response does not go below baseline
7.4.1 Unstable baseline
Some baseline drift is common in the first few cycles of an assay, and 3 to 5
start-up cycles are recommended to allow the assay to stabilize. Always check
the baseline in the reference and active sensorgrams separately: although a
drifting baseline may seem to be corrected by reference subtraction, it may
indicate inadequate assay optimization and complicate interpretation of the
results.
It is relevant to distinguish between baseline drift during an analysis cycle and
shifts in baseline response between cycles. The latter is most commonly caused
by regeneration characteristics, and can usually be ignored if the surface
response
time
1
2,3
4
6
5
5
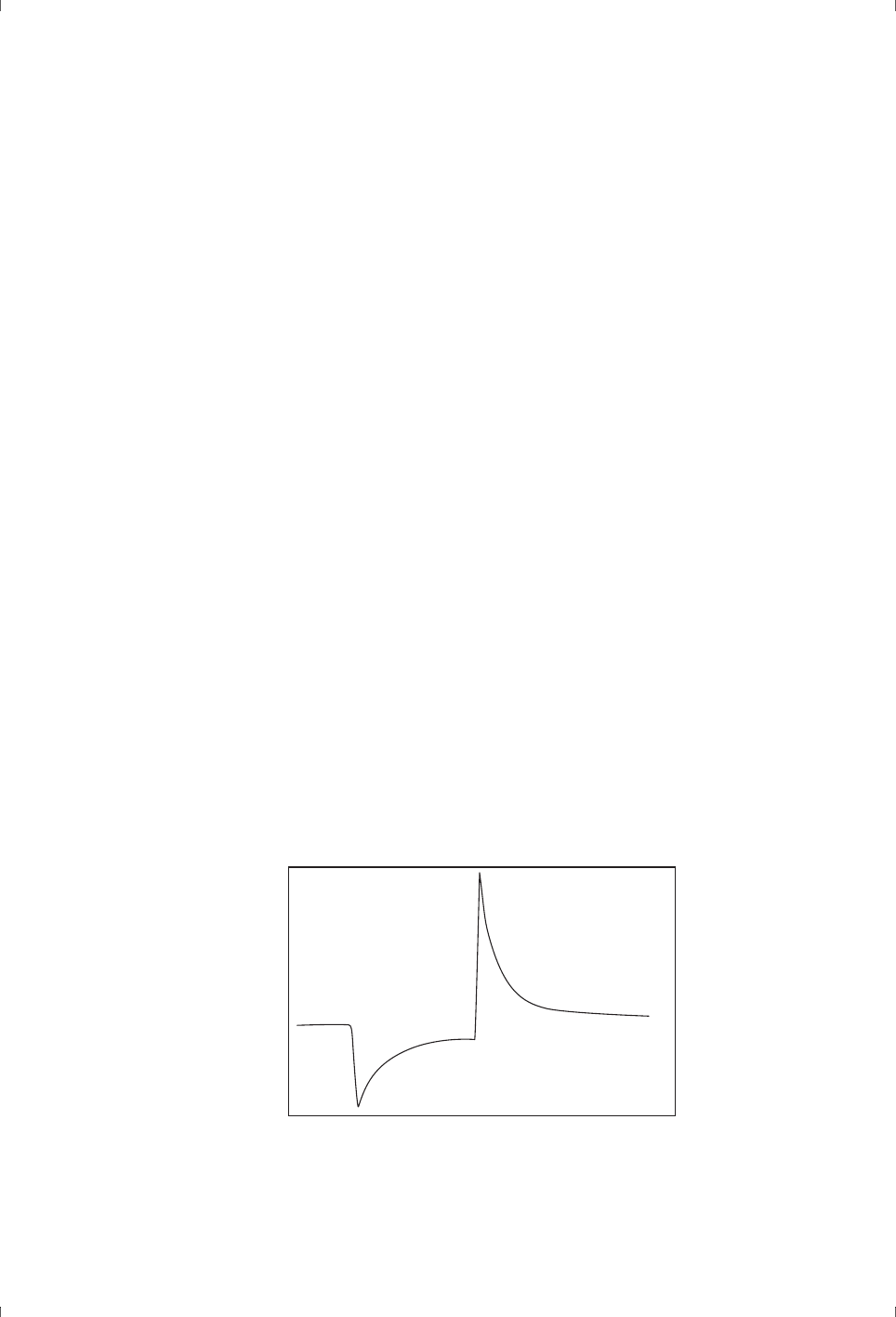
7 Troubleshooting assays
7.4 Unexpected sensorgram shapes
54 Biacore Assay Handbook 29-0194-00 Edition AA
binding capacity is not affected (as shown by reproducible binding levels for
control samples).
Baseline instability may also be caused by faults in the temperature control
system. If this occurs (indicated by a warning on the screen) call your
GE Healthcare service representative.
Baseline constantly increasing
• Make sure the running buffer is prepared from pure reagents using clean
glassware, and that the instrument flow system is thoroughly cleaned
after the previous experiment. This is particularly important for
applications using hydrophobic surfaces (Sensor Chip HPA and to a lesser
degree Sensor Chip L1).
Baseline constantly decreasing
• If you are using a capturing approach for attaching your ligand, check the
stability of the capturing interaction. For work with low molecular weight
analytes, you may be able to cross-link the ligand to the capturing
molecule after capture to stabilize the surface, but this results in
permanent attachment of the ligand. Cross-linking the ligand generally
reduces the binding activity for protein analytes.
• Downward drift of the baseline can also be the result of immobilizing a
multimeric ligand so that not all subunits are attached to the surface. If the
multimer dissociates, subunits will be lost, in most cases resulting in loss of
analyte binding capacity as well as baseline response. This behavior can
sometimes be prevented by crosslinking the ligand with EDC/NHS after it
has been attached to the surface, although the analyte binding activity
may be reduced.
7.4.2 Sample response below baseline
Figure 7-2. Sample response below baseline is usually be caused by a negative bulk
refractive index shift.
response
time

Biacore Assay Handbook 29-0194-00 Edition AA 55
Troubleshooting assays 7
Responses may fall below baseline during sample injection if the refractive
index of the sample is lower than that of running buffer, so that a negative bulk
refractive index shift is associated with the sample injection. In such cases, there
should be a corresponding positive shift at end of the injection: response levels
measured after the end of the injection will still be reliable.
Higher relative response in the reference flow cell than the active flow cell,
resulting from sample binding to the reference cell can also cause a shift below
baseline in the reference-subtracted sensorgram.
Both of these situations can usually be identified by examining the active and
reference sensorgrams separately.
7.4.3 “Humpbacked” sensorgrams
Sensorgrams with a “humpbacked” appearance during sample injection (where
the response reaches a maximum and then decreases, see Figure 3-4) are
relatively common when the detergent concentration in the running buffer is
below the critical micelle concentration (CMC). The recommended
concentration of Surfactant P20 is 0.05%. This detergent effect is usually
relatively small, and may go unnoticed in experiments with macromolecular
interactants where responses are in the range of several hundred RU and
above.
There are other possible causes of “humpbacked” sensorgrams, for example
initial binding of a large analyte that is then displaced by a slower but tighter
binding smaller molecule. You should be aware that this sensorgram
appearance does not always indicate a problem with the assay.
7.4.4 Anomalous response during buffer flow
Anomalous responses when only buffer is flowing through the system, including
both upward drift and response levels below baseline, may be caused by
contamination of the flow system with material from previous runs. Make sure
the instrument is maintained as described in the Instrument Handbook. If this
behavior is observed, clean the flow system thoroughly as instructed in the
Instrument Handbook, then allow the instrument to equilibrate with a low flow
of running buffer at least overnight.
7 Troubleshooting assays
7.4 Unexpected sensorgram shapes
56 Biacore Assay Handbook 29-0194-00 Edition AA
7.4.5 Response subtraction spikes
Figure 7-3. Reference subtraction spikes at the beginning and end of a blank (buffer)
injection.
Some overshoot at the start and end of the sample injection is normal in
reference-subtracted sensorgrams from systems with serially arranged flow
cells, particularly at low flow rates. This results from slight misalignment of the
reference and active sensorgrams together with a significant bulk refractive
index effect (Figure 7-3). This should not affect more than about 1 second of the
sensorgram. More serious overshoot may indicate malfunction of the sample
injection system, and requires attention from your GE Healthcare service
representative.
7.4.6 Regular disturbances
Disturbances (usually small) that occur repeatedly at the same time in each
cycle (Figure 7-4) are generally attributable to pump flow-fill operations.
Figure 7-4. Disturbances that occur at the same time in each cycle are attributable to
automatic aspects of instrument operation such as pump flow-fill.

Biacore Assay Handbook 29-0194-00 Edition AA 57
Analysis of kinetics and concentration measurements Appendix A
Appendix A Analysis of kinetics and concentration
measurements
A.1 Basic principles of kinetics and affinity
This section describes the basic principles for kinetics and affinity analysis of 1:1
interactions. Similar principles apply to other interaction models with
appropriately modified equations.
A.1.1 Kinetic rate equations
The kinetic evaluation procedure determines association and dissociation
constants by fitting the experimental data to a 1:1 interaction model between
analyte A and ligand B:
The net rate of complex formation during injection is given by
and the rate of dissociation after the end of the injection is
The concentration of complex formed is measured in RU by the SPR response: if
the total ligand concentration [B]
0
is also expressed in RU (as the maximum
analyte binding capacity), the rate equations can be written in terms of
response values instead of concentrations:
Because interaction occurs at the surface of the sensor chip, analyte must be
transferred laterally in the flow cell from the bulk solution to the surface before
interaction with ligand can take place. In a controlled flow system such as
Biacore, this transfer is a diffusion-limited process, referred to as mass transfer,
that can be described by a well-defined mathematical model. The analyte
where: k
a
is the association rate constant (M
-1
s
-1
)
and k
d
is the dissociation rate constant (s
-1
)
dAB
dt
---------------
k
a
AB k
d
AB–=
dAB
dt
---------------
k
d
AB–=
dR
dt
------
k
a
CR
max
k
a
Ck
d
+R–=

Appendix A Analysis of kinetics and concentration measurements
A.1 Basic principles of kinetics and affinity
58 Biacore Assay Handbook 29-0194-00 Edition AA
concentration available for interaction with the ligand is the concentration at
the sensor surface A
surface
, which is related to the concentration in the injected
sample A
bulk
by the equation:
where k
m
is the mass transfer coefficient, describing the diffusion-controlled
transfer from bulk solution to the surface. The transfer properties are the same
both to and from the surface, so that k
m
applies in both directions.
The mass transfer coefficient k
m
is a function of the flow rate, flow cell
dimensions and diffusion properties of the analyte molecule:
To minimize mass transport limitations in kinetic determinations, experiments
should be run at moderate to high flow rates (30 µl/min or more).
The mass transfer coefficient can be adjusted approximately for the molecular
weight of the analyte and for the conversion of surface concentration to RU to
give a term called the mass transfer constant k
t
:
A further modification of this expression gives the flow rate-independent
component of the mass transfer constant, referred to as tc:
For globular proteins with molecular weight of the order of 50,000 daltons,
typical values for tc are of the order of 10
8
RU·M
-1
s
-1
. Reported values that differ
greatly in order of magnitude (e.g. 10
12
or 10
14
) may indicate that the parameter
is not significant for the fitting (i.e. that the observed binding is not limited by
mass transfer).
where D is the diffusion coefficient of the analyte (m
2
s
-1
)
f is the volume flow rate of solution through the flow cell (m
3
s
-1
)
h, w, l are the flow cell dimensions (height, width, length in m)
k
m
0.98
D
h
---
23
f
0.3 w l
----------------------
13
=
k
t
k
m
MW 10
9
=
t
c
k
t
f
3
------
=

Biacore Assay Handbook 29-0194-00 Edition AA 59
Analysis of kinetics and concentration measurements Appendix A
Mass transfer is incorporated into the curve-fitting models by modifying the
concentration of analyte available for binding to ligand:
A.1.2 Steady-state affinity equations
At steady-state, the net rate of complex formation is zero:
so
Rearranging and setting R=R
eq
(the equilibrium response level) gives:
Setting k
a
/k
d
=K
A
(the equilibrium association constant) gives
The value of K
A
is obtained by fitting a plot of R
eq
against C to this equation.
K
D
is calculated as the inverse of K
A
(K
D
=1/K
A
).
A.1.3 Fitting procedure
The fitting procedure used to determine rate constants from experimental data
uses numerical integration methods with an iterative approximation algorithm
to find the best solution to the equations given above. The closeness of fit is
judged in terms of the chi-square value which describes the deviation between
the experimental and fitted curves:
where r
f
is the fitted value at a given point
r
x
is the experimental value at the same point
n is the number of data points
and p is the number of fitted parameters.
dR
dt
------
k
a
CR
max
k
a
Ck
d
+R– 0==
k
a
CR
max
k
a
Ck
d
+R=
R
eq
k
a
k
d
-----
C1+
k
a
k
d
-----
CR
max
=
R
eq
K
A
CR
max
K
A
C1+
-----------------------
=
chi-square
r
f
r
x
–
2
1
n
np–
----------------------------
=

Appendix A Analysis of kinetics and concentration measurements
A.1 Basic principles of kinetics and affinity
60 Biacore Assay Handbook 29-0194-00 Edition AA
The fitting algorithm seeks to minimize chi-square, and is judged to be complete
when the difference in chi-square values between successive iterations is
sufficiently small.
A.1.4 Assessing the fit
Kinetics
The rate constants reported by kinetic evaluation are determined in terms of a
1:1 interaction model, and it is important to realize that they are only valid in that
context. If the interaction mechanism is not a simple 1:1 binding, the fitted
curves will deviate to some extent from the experimental data and the reported
constants will not be a true representation of the interaction kinetics. The
apparent 1:1 binding constants can still be used for comparative studies of
observed binding rates, but it is important in reporting the values to emphasize
that they are empirical and not mechanistic constants.
There are two major tools for assessing the significance of the reported
constants: the closeness of fit between the fitted and experimental curves and
the statistical significance of the parameters.
Closeness of fit
Visual inspection of the residual plot or of the fitted curves overlaid on the
experimental data gives an indication of the closeness of fit. Ideally, the
residuals will scatter randomly around zero over a range that corresponds to
the short-term noise in the detection system (approximately 1 to 2 RU).
Deviations from ideal fitting appear as systematic variations in the residuals,
imparting a non-linear shape to the residual plot. Judge the residual range and
shape in proportion to the response ranges in the experimental sensorgrams
and in relation the goal of the investigation.
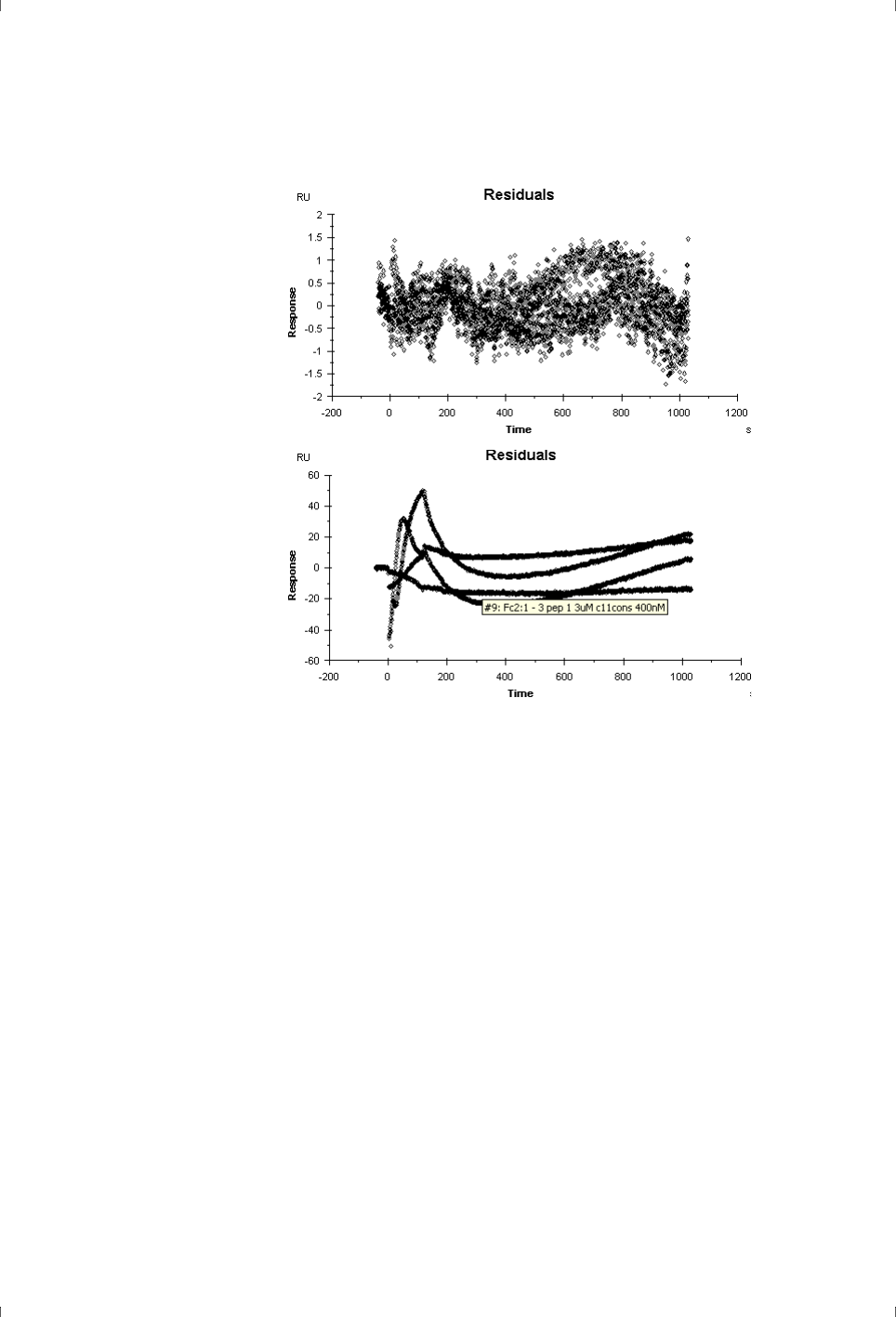
Biacore Assay Handbook 29-0194-00 Edition AA 61
Analysis of kinetics and concentration measurements Appendix A
Figure A-1. Residual plots from a good fit (top) and a poor fit (bottom). Note the different
response scales in the two plots (top ±2 RU, bottom ±60 RU).
The chi-square value is a quantitative measure of the closeness of fit, and in an
ideal situation will approximate to the square of the short-term noise level. It is
however difficult to recommend absolute values for acceptance limits for chi-
square: the values need to be considered from case to case in combination with
assessment of the shape of the residuals (chi-square is related to the overall
range of the residuals but is not affected by the shape of the residual curve).
Significance of parameters
Fitting experimental data to a mathematical interaction model will return values
for all parameters in the model, regardless of whether they are significant or
not. As an example, sensorgrams that are completely limited by mass transfer
will give values for the kinetic rate constants even though the experimental data
does not contain any kinetic information.
The significance of the parameter values returned by the fitting procedure is
indicated by the standard error or T-value, which broadly speaking represents
the degree to which the value can be varied without significantly affecting the
closeness of fit. The T-value is the standard error divided by the parameter
value. A high standard error or low T-value indicates that the fitting is insensitive

Appendix A Analysis of kinetics and concentration measurements
A.2 Interaction models for kinetics
62 Biacore Assay Handbook 29-0194-00 Edition AA
to changes in the parameter value: in other words, the value returned for the
parameter has little significance.
It is important to examine the significance of values calculated by the software,
to avoid the trap of reporting “constants” that have no significance. In the
example of completely mass transfer limited interactions, the rate constants
will have a high standard error (low T-value), since it does not matter for the
fitting result what value will be returned.
Note: Even if the significance of a parameter is low, so that any value within a
wide range will give the same fit to the experimental data, the fitting
algorithm will always return the same value from the same starting data.
Consistency of returned values is not an indication of significance.
For the 1:1 binding model, an additional indicator of the parameter significance
is the U-value. This is a parameter that represents the uniqueness of the
calculated rate constants and R
max
, determined by testing the dependence of
fitting on correlated variations between selected variables. Lower values
indicate greater confidence in the results. A high value (above about 25)
indicates that the reported kinetic constants contain no useful information.
Affinity
To obtain a robust fit to a plot of R
eq
against C for affinity determination, it is
important that the range of analyte concentration is wide enough to reveal the
curvature of the plot in full. In particular, if the highest concentration is too low
in relation to the calculated K
D
value, the reported values will be uncertain even
if the fit is apparently good and the chi-square value is low.
As a general guideline, the fit will be reliable if the highest analyte concentration
is at least twice the calculated K
D
value. If the highest analyte concentration is
less than 2K
D
, you should treat the reported K
D
with caution.
A.2 Interaction models for kinetics
This section gives a brief description of the standard kinetic interaction models
provided with Biacore systems.
A.2.1 1:1 binding
This interaction model describes one molecule of analyte A binding to one
molecule of ligand B:

Biacore Assay Handbook 29-0194-00 Edition AA 63
Analysis of kinetics and concentration measurements Appendix A
A.2.2 1:1 dissociation
This model fits the dissociation phase only to an exponential model. The
dissociation rate is independent of the analyte concentration in the injected
sample, so that dissociation rate constants can be obtained for samples with
unknown concentration.
This model does not include a mass transfer coefficient. If mass transfer
limitations are significant, dissociated analyte will be washed away from the
surface with reduced efficiency and the dissociation rate constant will be
underestimated.
A.2.3 Bivalent analyte binding
The bivalent analyte binding model describes the interaction of monovalent
ligand with analyte molecules that carry two identical and independent binding
sites.
Once binding has occurred at the first analyte site, binding at the second site is
facilitated by the proximity of analyte and ligand. Similarly, analyte molecules
that are attached at both sites are not released from the surface until
dissociation occurs at both sites, so the observed dissociation rate is much
slower than that seen for a single site. The resulting enhanced binding is often
called avidity to distinguish it from single site binding affinity.
Note: The association rate constant for interaction at the second site is reported
in units of RU
-1
s
-1
. This is because both the interacting components are
present on the surface and are measured in RU, not in true
concentrations.
where k
a
is the association rate constant
and k
d
is the dissociation rate constant
where k
a1
and k
d1
are the association and dissociation rate constants for the
first site
and k
a2
and k
d2
are the association and dissociation rate constants for the
second site

Appendix A Analysis of kinetics and concentration measurements
A.3 Concentration measurements
64 Biacore Assay Handbook 29-0194-00 Edition AA
A.2.4 Heterogeneous ligand
This model describes interaction of one analyte with two independent ligands
(or ligand sites) on the surface. The observed binding is the sum of the
interaction with the two ligands.
A.2.5 Interaction models for affinity
1:1 steady state binding
The Affinity evaluation item described in Chapter 12 fits a plot of response
against concentration to the equation (Section A.1.2)
The parameters K
A
, R
max
and offset are fitted as variables. The affinity is
reported as a K
D
value, which is the inverse of K
A
. This function gives reliable
fitting if the highest analyte concentration used is at least twice the K
D
value.
A.3 Concentration measurements
A.3.1 Calibration curve fitting
Calibration curves for concentration measurement are fitted to either linear or
4-parameter equations. Custom equations for calibration curves can be defined
in some Biacore systems.
where k
a1
and k
d1
are the association and dissociation rate constants for one
ligand
and k
a2
and k
d2
are the association and dissociation rate constants for the
other ligand
where offset is a constant term that gives the intercept of the fitted curve on
the y-axis. Without this term the fitted curve is forced to go through the
origin of the plot.
R
eq
K
A
CR
max
K
A
C1+
-----------------------
offset+=

Biacore Assay Handbook 29-0194-00 Edition AA 65
Analysis of kinetics and concentration measurements Appendix A
For linear fitting, the points are fitted to the equation
y = slope * x + intercept
The equation for a 4-parameter fit is
The closeness of fit is reported for linear fitting as the coefficient of
determination R
2
and for 4-parameter fitting as the chi-square value.
A.3.2 Calibration trends
Some Biacore systems support calibration trends to compensate for drift in the
calibration throughout the course of the assay. With this approach, calibration
curves are measured at the start and end of the assay and at intervals, and an
individual calibration curve is constructed for each sample analysis cycle by
interpolating between the measured calibration curves.
A.3.3 Calibration-free concentration analysis
Evaluation of calibration-free concentration relies on fitting the data to a model
for 1:1 interaction (see Section A.1) where the mass transfer coefficient is
provided (through calculation from the diffusion coefficient and flow cell
characteristics) and the analyte concentration is evaluated as a global variable.
This model is equivalent in terms of the interaction description to the model for
evaluation of 1:1 kinetics.
where y and x are the plot coordinates
R
hi
and R
lo
are fitting parameters that correspond to the maximum and
minimum response levels respectively
A
1
and A
2
are additional fitting parameters
y R
hi
R
hi
R
lo
–
1
x
A
1
----- -
A
2
+
--------------------------
–=

Appendix A Analysis of kinetics and concentration measurements
A.3 Concentration measurements
66 Biacore Assay Handbook 29-0194-00 Edition AA
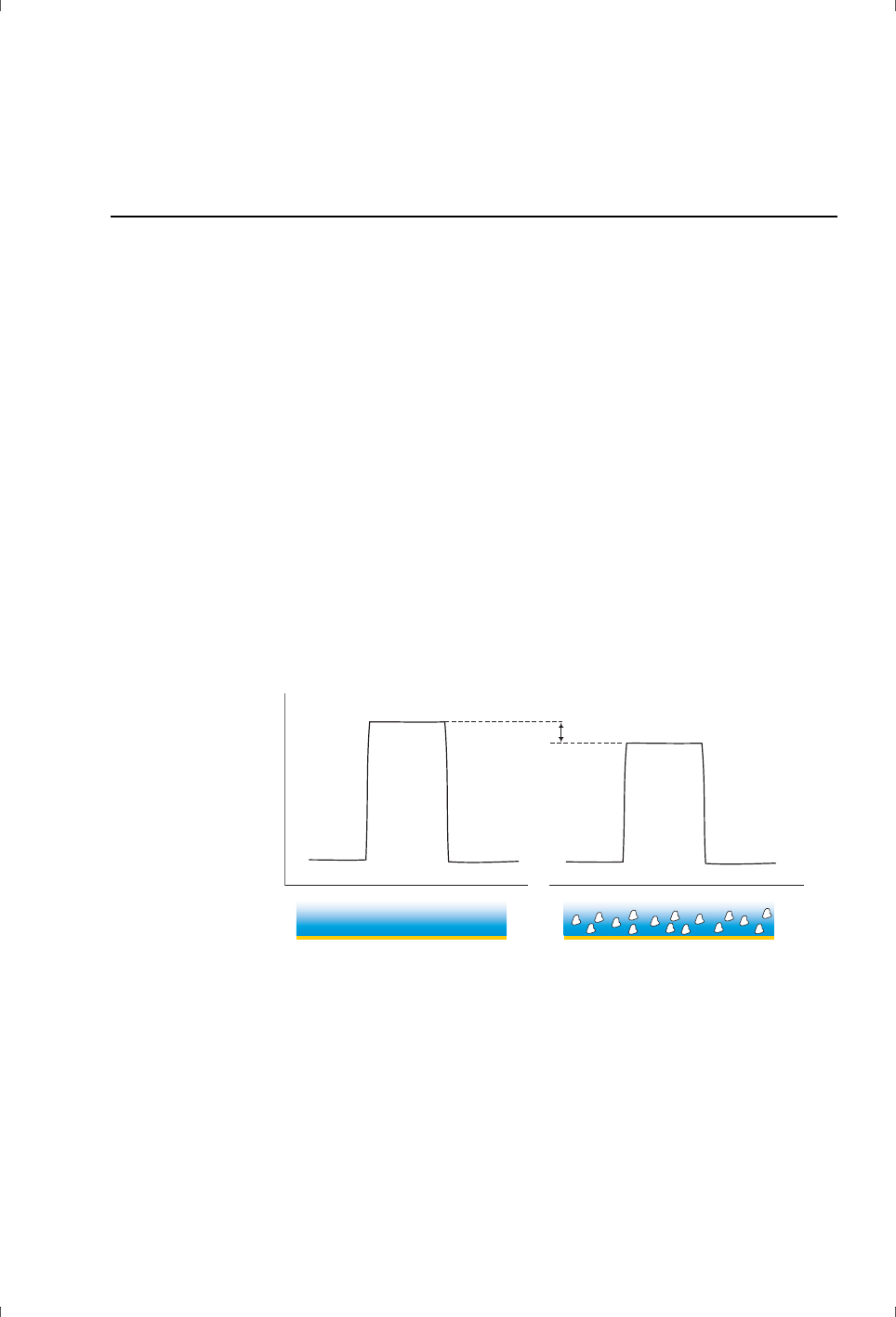
Biacore Assay Handbook 29-0194-00 Edition AA 67
Solvent correction principles and practice Appendix B
Appendix B Solvent correction principles and practice
B.1 Introduction
Solvent correction refers to the procedures used to adjust measured responses
for the effects of varying concentrations of organic solvents. These procedures
are relevant primarily for work with small organic analytes that require inclusion
of organic solvents (commonly dimethyl sulfoxide, DMSO) in the buffer to
maintain analyte solubility.
Simple subtraction of a reference response is not sufficiently accurate for these
purposes when differences in bulk shifts between samples can be of the same
order of magnitude as (or larger than) the expected response from analyte
binding. One reason for this is that the volume of solution accessible to sample
will differ on the reference and active surfaces unless the two surfaces have the
same attached protein concentration. As a result of the volume excluded by
ligand molecules, the bulk shift will be smaller on the active surface than the
reference surface (Figure B-1).
There are other factors in addition to excluded volume involved in solvent
correction that are outside the scope of this discussion, so that solvent
correction is essentially an empirical procedure.
Figure B-1. Bulk solution is excluded from the volume occupied by ligand molecules on the
active surface, so the contribution of bulk solution to the relative response is smaller than
on the reference surface.
B.2 Requirement for solvent correction
Solvent correction is necessary under a combination of three circumstances,
commonly met in work with low molecular weight analytes such as drug
candidates:
Response on
reference surface
Response on
active surface
Excluded volume effect
Active surface
Reference surface
Appendix B Solvent correction principles and practice
B.3 Solvent correction principles
68 Biacore Assay Handbook 29-0194-00 Edition AA
• the expected analyte responses are low,
• the ligand is a macromolecule immobilized at a high density on the
surface,
• the bulk response is high in relation to the measured binding response.
In drug discovery and development work, the analytes are often small
molecules which give correspondingly low response values (typically of the
order of 10-50 RU or less). High levels of immobilized ligands (several thousand
RU) are used to maximize the analyte response, enhancing the excluded volume
effect described above. Samples generally include DMSO to maintain solubility,
resulting in high bulk responses. A difference of 1% (percentage points) in DMSO
concentration corresponds to a difference of about 1200 RU in response, so that
small variations in DMSO concentration, unavoidable in the preparation of
diverse samples, easily lead to variations in bulk response of the same order of
magnitude as the expected sample responses.
B.3 Solvent correction principles
Solvent correction values are determined by injecting blank samples containing
a range of DMSO concentrations and plotting the reference-subtracted
response from the active surface against the relative response from the
reference surface. This creates a calibration curve, referred to as the solvent
correction curve, for the solvent effect as a function of the reference response.
Each sample measurement can then be adjusted for the solvent effect on the
basis of the response on the reference surface (see Figure B-2).
Figure B-2. The principle of solvent correction. 1. The sensorgram from the reference
surface shows a bulk displacement (-150 RU in the illustration) during sample injection
because the sample and running buffer are not exactly matched. 2. From the solvent
correction curve, a displacement of –150 RU in the reference sensorgram corresponds to
a solvent effect of +5 RU in the reference-subtracted sensorgram. 3. The reference-
subtracted sensorgram is corrected for the solvent error. This procedure is applied to
every point during sample injection.
Solvent correction is applied only to response levels during sample injection,
since the correction adjusts for differences in bulk response between different
samples.

Biacore Assay Handbook 29-0194-00 Edition AA 69
Solvent correction principles and practice Appendix B
B.4 Preparing solutions for solvent correction
It is important to match samples and running buffer as closely as possible with
respect to refractive index to minimize bulk refractive index effects, so that
solvent correction values are kept small. It is particularly important to strive for
exactly the same DMSO concentration in samples and running buffer and to
minimize variations in DMSO content between samples. Bear the following in
mind when preparing samples for assays where solvent correction is necessary:
• Use carefully designed buffer preparation and sample dilution protocols to
ensure consistent DMSO concentrations. Samples that are stored in DMSO
should be diluted in such a way as to give the same final DMSO
concentration as in the running buffer.
• Cover the sample plates with foil directly after preparation to avoid
changes in DMSO concentration resulting from evaporation and
absorption of water from the air. Remember that DMSO is hygroscopic.
• If you are using a laboratory robot to prepare samples, consider running a
test with blank samples to establish the range of variability of DMSO
concentrations in the samples. Run solvent correction cycles that cover
the expected range of variability.
Laboratory protocols for preparation of samples and solvent correction
solutions may be found on www.gelifesciences.com/biacore or obtained from
GE Healthcare.
B.5 Assessing solvent correction procedures
Solvent correction curves often show a negative slope with a slight curvature
(Figure B-3, left panel), but the slope and shape is not predictable and curves
with positive slope or even with a maximum or minimum within the curve may
be acceptable.
It is however important that the solvent correction curves fit the experimental
points closely. Poor fitting of the curves to the experimental points (Figure B-3,
center panel) can introduce uncertainties in the solvent correction factors that
are the same order of magnitude as the factors themselves, potentially
introducing additional errors in the sample measurements.
Sample responses that lie outside the range of the solvent correction curves
(Figure B-3, right panel) cannot be corrected and will be omitted from evaluation
of solvent corrected data. Solvent correction curves may be extrapolated in the
software, but this option should only be used for small extensions of the solvent
correction range (less than 10% of the measured range) since the curve shape
is not predictable.
Appendix B Solvent correction principles and practice
B.5 Assessing solvent correction procedures
70 Biacore Assay Handbook 29-0194-00 Edition AA
Figure B-3. Some examples of solvent correction curves.
Left: Acceptable curve, with close fitting to the experimental points and covering the
sample response range well.
Center: The experimental points scatter widely around the fitted solvent correction curve.
Badly fitted curves should be treated with caution.
Right: The solvent correction curve does not cover the whole sample response range.
Sample responses outside the range of the curve will not be corrected.

Biacore Assay Handbook 29-0194-00 Edition AA 1
Numerics
1:1 binding 57
, 62
1:1 dissociation 63
2-over-2 kinetics 39
A
absolute response 8
acidification-neutralization 38
activity of surface-attached ligand 22
ADAs, see anti-drug antibodies
affinity 6
, 39
affinity analysis 11
affinity constant 8
, 10
affinity evaluation
interpreting the fit 62
affinity in solution 12
affinity model 64
affinity screen 35
amine coupling 20
analysis conditions
for antibody screening 36
for small molecule screening 33
analyte 7
analyte binding capacity 22
analyte concentration 13
analyte concentration range 42
antibody dissociation rate 47
antibody ranking 47
antibody screening 31
, 35
antibody specificity 14
, 37
anti-drug antibodies 31
, 38
association phase 10
association rate constant 57
, 63, 64
attaching the ligand 19
avidity 63
B
baseline 52
baseline drift 53
Biacore systems 6
Biacore terminology 7
binding level screen 35
biopharmaceutical screening 31
biotinylated ligands 50
bivalent analyte 63
blocking antibody 45
buffer composition 27
buffer exchange 28
buffer matching 41
buffer substances 23
buffers
for antibody screening 36
for kinetics and affinity 41
for small molecule screening 32
bulk effects 41
bulk refractive index 25
bulk shift 8
, 26
C
caibration-free concentration analysis 13
calibrated assays 13
calibration curve 14
calibration-free concentration analysis 14
, 42, 44
capture 19
, 21
capturing molecule 7
carry-over 34
CFCA, see calibration-free concentration analysis
change in surface concentration 9
chemical modification 21
chi-square 59
, 61, 65
clean screen 35
closeness of fit 59
, 60
coefficient of determination 65
competition assays 13
complex sample matrices 31
complex samples 9
concentration measurement 6
, 13
coupling chemistry 19
covalent immobilization 19
critical micelle concentration 24
, 55
crowding 40
cut-off 34
D
degassing buffers 25
desalting 28
detecting binding partners 9
detecting molecule 7
, 13
detection principle 5
detergent 24
, 33, 55
dextran matrix 17
dialysis 28
diffusion coefficient 14
, 58
diffusion properties 14
diffusion-limited binding 13
diluting from stock solution 28
, 42
dilution protocol 69
dilution series 28
dimethyl sulfoxide 25
, 32
direct binding assay 13
dissociation fit 63
dissociation phase 10
dissociation rate constant 57
, 63, 64
DMSO, see dimethyl sulfoxide
drug discovery 5
E
EDC/NHS 22

Biacore Assay Handbook 29-0194-00 Edition AA 2
EDTA 25
enhancement 13
enzyme kinetics 10
epitope mapping 5
, 14, 45
equilibrium constant 8
equilibrium response level 59
evaluation of epitope mapping 46
expected response 22
extrapolating solvent correction 69
F
fitting procedure 59
flow rate 58
four-parameter fit 65
fragment screening 32
, 35
H
HEPES 23
heterogeneous ligand 64
high affinity capture 19
, 21
high analyte binding 51
histidine-tagged ligands 19
, 50
humpbacked sensorgrams 55
I
immobilization 7
, 19
immobilization chemistry 20
immobilization pH 49
immobilization sensorgram 23
immobilized ligand for kinetics and affinity 40
immunogenicity 5
, 31, 38
indirect assays 13
information-rich screening 32
inhibition 13
interaction kinetics 6
, 10
interaction mechanism 10
interaction models 10
, 62
interaction specificity 5
interpreting the fit 60
ionic strength 23
, 49
K
K
A
, K
D
59
k
a
, k
d
57
kinetic evaluation
interpreting the fit 60
kinetic interaction models 62
kinetics 6
, 39
k
m
58
k
t
58
L
label-free technology 5
laboratory robot 69
ligand 7
ligand concentration 19
linear fit 65
lipid monolayers 18
lipid vesicles 18
LMW screening, see small molecule screening
low analyte capacity 51
M
maintenance tools 24
mapping binding sites 14
mass transfer constant 58
mass transfer limitation 40
mass transport 57
mass transport coefficient 58
matching sample and running buffer 26
micro-spin columns 28
molar response 9
molecular weight dependence 5
multi-cycle kinetics 11
, 39
N
negative controls 34
nitrilotriacetic acid 19
non-saturatable binding 52
non-specific binding 23
, 25, 51
NSB Reducer 25
, 52
O
offset 64
organic solvents 25
, 32
P
pair-wise binding 15
, 45
pair-wise matrix 46
parameter significance 61
peptide inhibition 15
, 45, 48
permanent capture 19
phosphate buffers 23
plasma samples 26
pre-concentration 19
promiscuous binding 32
R
rate constants 8
, 10
rate equations 57
ready-to-use buffers 23
reference subtraction 27
reference surface 40
, 51
refractive index changes 5
refractive index differences 8
regeneration 8
, 19
relative interaction kinetics 40
relative response 8
replicates 29
report points 8
, 27

Biacore Assay Handbook 29-0194-00 Edition AA 3
R
eq
59
residuals 60
resonance unit 7
response 7
response below baseline 54
response enhancement 13
response levels 21
response overshoot 53
, 56
reversible capture 19
R
max
22
running buffer 7
S
sample concentrations for kinetics and affinity 42
sample injection 7
sample preparation 32
, 36, 41
sandwich assays 13
Scatchard plots 12
screening 5
, 9
sensor chip 17
sensor surfaces 17
sensorgram 8
sensorgram disturbances 56
sensorgram shape 52
serum samples 26
significance 61
single-cycle kinetics 11
, 39
small molecule screening 31
, 32
sodium azide 25
, 50
solid samples 28
solution competition 13
solvent correction 25
, 27, 33
range 69
solvent correction curves
typical appearance 69
SPR, see surface plasmon resonance
standard error 61
start-up cycles 53
steady state affinity 12
steady state binding 59
, 64
sticky compounds 34
streptavidin 19
subtraction spikes 56
surface activation 22
surface competition 13
surface plasmon resonance 5
Surfactant P20 24
T
target molecule 7
, 32
t
c
58
terminology 7
thiol coupling 20
Tris buffer 23
, 50
troubleshooting 49
T-value 61
Tween 24
U
uniqueness 62
units for k
a2
63
unwanted binding 52
U-value 62
V
vials and microplates 30

4 Biacore Assay Handbook 29-0194-00 Edition AA


imagination at work
29-0194-00 AA 05/2012
For local office contact information visit
www.gelifesciences.com/contact
GE Healthcare Bio-Sciences AB
Björkgatan 30
751 84 Uppsala
Sweden
www.gelifesciences.com/biacore
GE, imagination at work and GE monogram are trademarks of General
Electric Company.
Biacore is a trademark of GE Healthcare companies.
Tween is a trademark of ICI Americas Inc.
© 2012 General Electric Company—All rights reserved.
First published May 2012
All goods and services are sold subject to the terms and conditions of sale
of the company within GE Healthcare which supplies them. A copy of these
terms and conditions is available on request. Contact your local GE
Healthcare representative for the most current information.
GE Healthcare UK Ltd
Amersham Place, Little Chalfont, Buckinghamshire, HP7 9NA, UK
GE Healthcare Bio-Sciences Corp
800 Centennial Avenue, P.O. Box 1327, Piscataway, NJ 08855-1327, USA
GE Healthcare Europe GmbH
Munzinger Strasse 5, D-79111 Freiburg, Germany
GE Healthcare Bio-Sciences KK
Sanken Bldg. 3-25-1, Hyakunincho, Shinjuku-ku, Tokyo 169-0073, Japan
